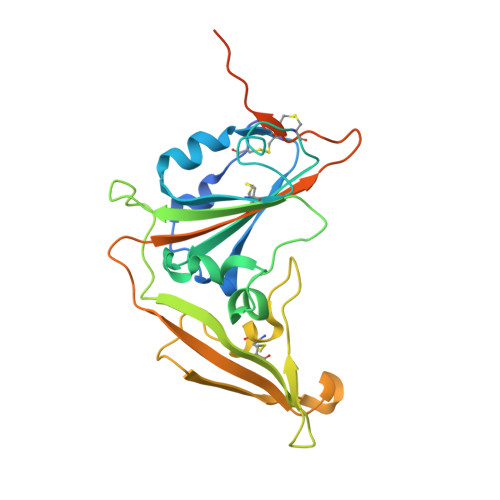
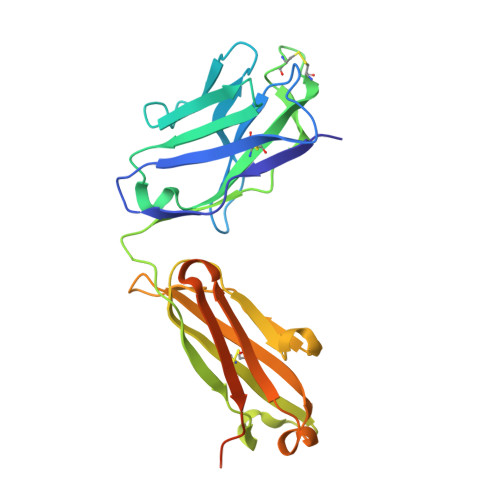
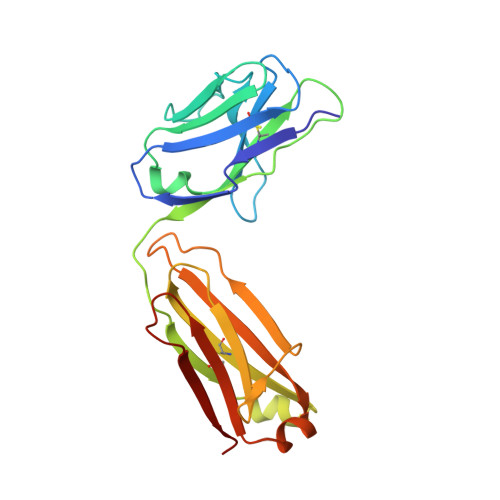
Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody.
Ying, T., Prabakaran, P., Du, L., Shi, W., Feng, Y., Wang, Y., Wang, L., Li, W., Jiang, S., Dimitrov, D.S., Zhou, T.(2015) Nat Commun 6: 8223-8223
- PubMed: 26370782
- DOI: https://doi.org/10.1038/ncomms9223
- Primary Citation of Related Structures:
4XAK - PubMed Abstract:
The MERS-CoV is an emerging virus, which already infected more than 1,300 humans with high (∼36%) mortality. Here, we show that m336, an exceptionally potent human anti-MERS-CoV antibody, is almost germline with only one somatic mutation in the heavy chain. The structure of Fab m336 in complex with the MERS-CoV receptor-binding domain reveals that its IGHV1-69-derived heavy chain provides more than 85% binding surface and that its epitope almost completely overlaps with the receptor-binding site. Analysis of antibodies from 69 healthy humans suggests an important role of the V(D)J recombination-generated junctional and allele-specific residues for achieving high affinity of binding at such low levels of somatic hypermutation. Our results also have important implications for development of vaccine immunogens based on the newly identified m336 epitope as well as for elucidation of mechanisms of neutralization by m336-like antibodies and their elicitation in vivo.
- Key Laboratory of Medical Molecular Virology of Ministries of Education and Health, Shanghai Medical College, Fudan University, Shanghai 200032, China.
Organizational Affiliation: