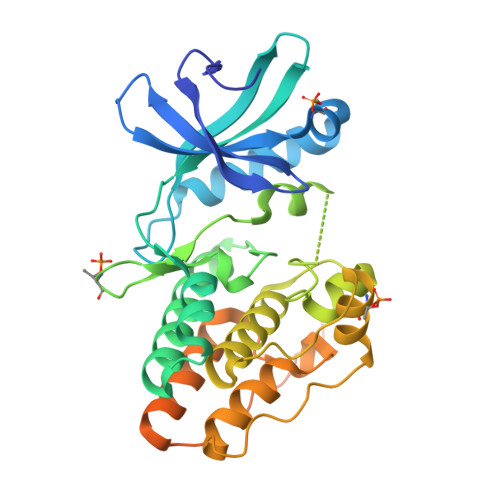
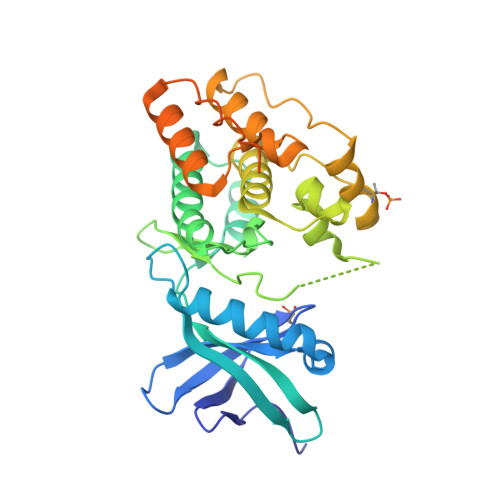
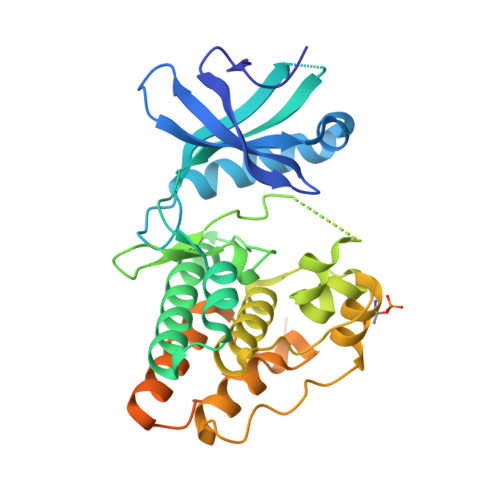
The crystal structure of the catalytic domain of the ser/thr kinase PknA from M. tuberculosis shows an Src-like autoinhibited conformation.
Wagner, T., Alexandre, M., Duran, R., Barilone, N., Wehenkel, A., Alzari, P.M., Bellinzoni, M.(2015) Proteins 83: 982-988
- PubMed: 25586004
- DOI: https://doi.org/10.1002/prot.24754
- Primary Citation of Related Structures:
4X3F - PubMed Abstract:
Signal transduction mediated by Ser/Thr phosphorylation in Mycobacterium tuberculosis has been intensively studied in the last years, as its genome harbors eleven genes coding for eukaryotic-like Ser/Thr kinases. Here we describe the crystal structure and the autophosphorylation sites of the catalytic domain of PknA, one of two protein kinases essential for pathogen's survival. The structure of the ligand-free kinase domain shows an auto-inhibited conformation similar to that observed in human Tyr kinases of the Src-family. These results reinforce the high conservation of structural hallmarks and regulation mechanisms between prokaryotic and eukaryotic protein kinases.
- Institut Pasteur, Unité De Microbiologie Structurale, Paris, 75724, France; CNRS UMR 3528, Paris, 75724, France; Université Paris Diderot, Sorbonne Paris Cité, Microbiologie Structurale, Paris, 75724, France.
Organizational Affiliation: