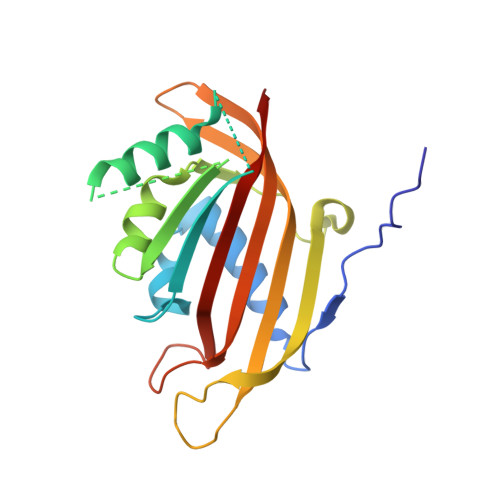
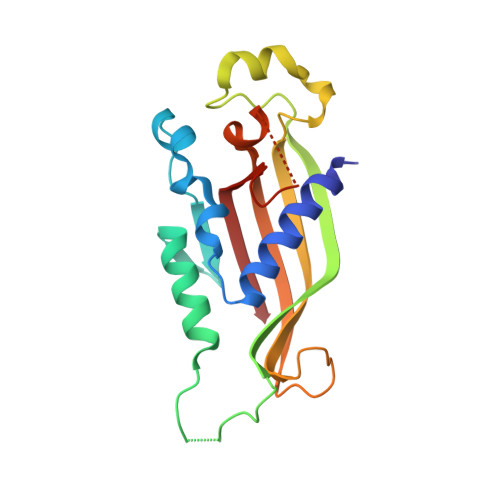
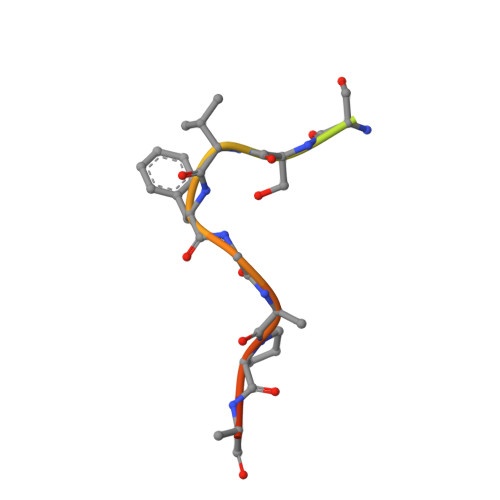
Structural Characterization of the Chaetomium thermophilum TREX-2 Complex and its Interaction with the mRNA Nuclear Export Factor Mex67:Mtr2.
Dimitrova, L., Valkov, E., Aibara, S., Flemming, D., McLaughlin, S.H., Hurt, E., Stewart, M.(2015) Structure 23: 1246-1257
- PubMed: 26051714
- DOI: https://doi.org/10.1016/j.str.2015.05.002
- Primary Citation of Related Structures:
4WPX, 4X2H, 4X2O - PubMed Abstract:
The TREX-2 complex integrates mRNA nuclear export into the gene expression pathway and is based on a Sac3 scaffold to which Thp1, Sem1, Sus1, and Cdc31 bind. TREX-2 also binds the mRNA nuclear export factor, Mex67:Mtr2, through the Sac3 N-terminal region (Sac3N). Here, we characterize Chaetomium thermophilum TREX-2, show that the in vitro reconstituted complex has an annular structure, and define the structural basis for interactions between Sac3, Sus1, Cdc31, and Mex67:Mtr2. Crystal structures show that the binding of C. thermophilum Sac3N to the Mex67 NTF2-like domain (Mex67(NTF2L)) is mediated primarily through phenylalanine residues present in a series of repeating sequence motifs that resemble those seen in many nucleoporins, and Mlp1 also binds Mex67:Mtr2 using a similar motif. Deletion of Sac3N generated growth and mRNA export defects in Saccharomyces cerevisiae, and we propose TREX-2 and Mlp1 function to facilitate export by concentrating mature messenger ribonucleoparticles at the nuclear pore entrance.
- Medical Research Council Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge Biomedical Campus, Cambridge CB2 0QH, UK; Biochemie-Zentrum der Universität Heidelberg, INF328, 69120 Heidelberg, Germany.
Organizational Affiliation: