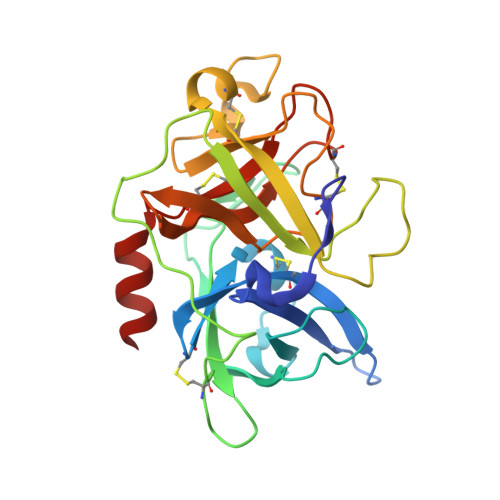
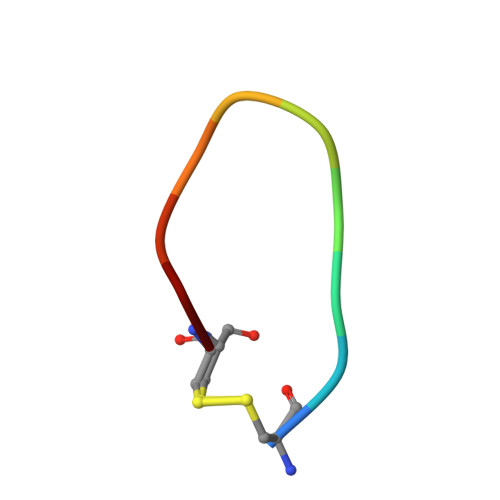
Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Jiang, L., Zhao, B., Xu, P., Srensen, H.P., Jensen, J.K., Christensen, A., Hosseini, M., Nielsen, N.C., Jensen, K.J., Andreasen, P.A., Huang, M.(2015) Int J Biochem Cell Biol 62: 88-92
- PubMed: 25744057
- DOI: https://doi.org/10.1016/j.biocel.2015.02.016
- Primary Citation of Related Structures:
4X0W, 4X1P - PubMed Abstract:
Two isomeric piperidine derivatives (meta and para isomers) were used as arginine mimics in the P1 position of a cyclic peptidic inhibitor (CPAYSRYLDC) of urokinase-type plasminogen activator. The two resulting cyclic peptides showed vastly different affinities (∼70 fold) to the target enzyme. X-ray crystal structure analysis showed that the two P1 residues were inserted into the S1 specificity pocket in indistinguishable manners. However, the rest of the peptides bound in entirely different ways on the surface of the enzyme, and the two peptides have different conformations, despite the highly similar sequence. These results demonstrate how the subtle difference in P1 residue can dictate the exosite interactions and the potencies of peptidic inhibitors, and highlight the importance of the P1 residue for protease inhibition. This study provides important information for the development of peptidic agents for pharmacological intervention.
- Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou 350002, China(1); Danish-Chinese Centre for Proteases and Cancer, China.
Organizational Affiliation: