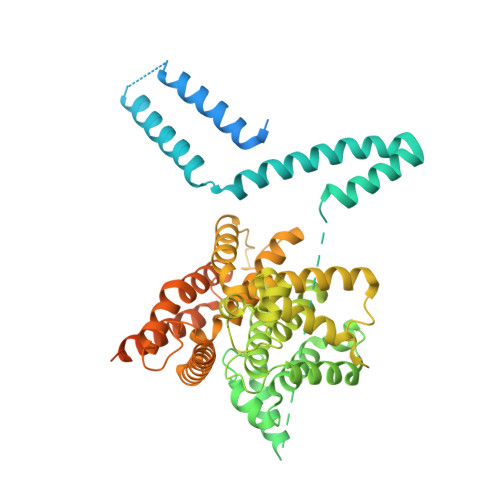
Engineered stabilization and structural analysis of the autoinhibited conformation of PDE4.
Cedervall, P., Aulabaugh, A., Geoghegan, K.F., McLellan, T.J., Pandit, J.(2015) Proc Natl Acad Sci U S A 112: E1414-E1422
- PubMed: 25775568
- DOI: https://doi.org/10.1073/pnas.1419906112
- Primary Citation of Related Structures:
4WZI, 4X0F - PubMed Abstract:
Phosphodiesterase 4 (PDE4) is an essential contributor to intracellular signaling and an important drug target. The four members of this enzyme family (PDE4A to -D) are functional dimers in which each subunit contains two upstream conserved regions (UCR), UCR1 and -2, which precede the C-terminal catalytic domain. Alternative promoters, transcriptional start sites, and mRNA splicing lead to the existence of over 25 variants of PDE4, broadly classified as long, short, and supershort forms. We report the X-ray crystal structure of long form PDE4B containing UCR1, UCR2, and the catalytic domain, crystallized as a dimer in which a disulfide bond cross-links cysteines engineered into UCR2 and the catalytic domain. Biochemical and mass spectrometric analyses showed that the UCR2-catalytic domain interaction occurs in trans, and established that this interaction regulates the catalytic activity of PDE4. By elucidating the key structural determinants of dimerization, we show that only long forms of PDE4 can be regulated by this mechanism. The results also provide a structural basis for the long-standing observation of high- and low-affinity binding sites for the prototypic inhibitor rolipram.
- Structural Biology and Biophysics Group, Worldwide Research and Development, Pfizer, Inc., Groton, CT 06340.
Organizational Affiliation: