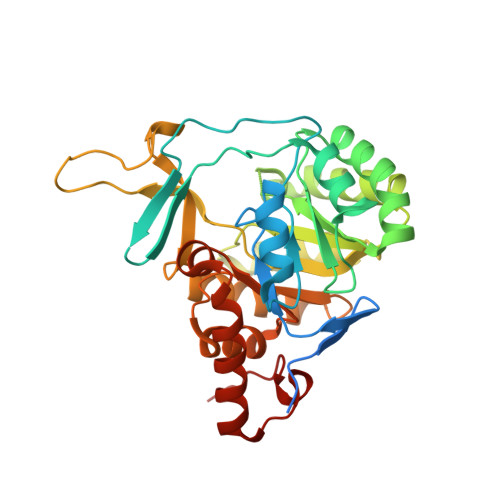
Recombinant production, crystallization and crystal structure determination of dihydroorotate dehydrogenase from Leishmania (Viannia) braziliensis.
Reis, R.A., Lorenzato, E., Silva, V.C., Nonato, M.C.(2015) Acta Crystallogr F Struct Biol Commun 71: 547-552
- PubMed: 25945707
- DOI: https://doi.org/10.1107/S2053230X15000886
- Primary Citation of Related Structures:
4WZH - PubMed Abstract:
The enzyme dihydroorotate dehydrogenase (DHODH) is a flavoenzyme that catalyses the oxidation of dihydroorotate to orotate in the de novo pyrimidine-biosynthesis pathway. In this study, a reproducible protocol for the heterologous expression of active dihydroorotate dehydrogenase from Leishmania (Viannia) braziliensis (LbDHODH) was developed and its crystal structure was determined at 2.12 Å resolution. L. (V.) braziliensis is the species responsible for the mucosal form of leishmaniasis, a neglected disease for which no cure or effective therapy is available. Analyses of sequence, structural and kinetic features classify LbDHODH as a member of the class 1A DHODHs and reveal a very high degree of structural conservation with the previously reported structures of orthologous trypanosomatid enzymes. The relevance of nucleotide-biosynthetic pathways for cell metabolism together with structural and functional differences from the respective host enzyme suggests that inhibition of LbDHODH could be exploited for antileishmanicidal drug development. The present work provides the framework for further integrated in vitro, in silico and in vivo studies as a new tool to evaluate DHODH as a drug target against trypanosomatid-related diseases.
- Departamento de Física e Química, Faculdade de Ciências Farmacêuticas de Ribeirão Preto, Avenida Café S/N, 14040-903 Ribeirão Preto-SP, Brazil.
Organizational Affiliation: