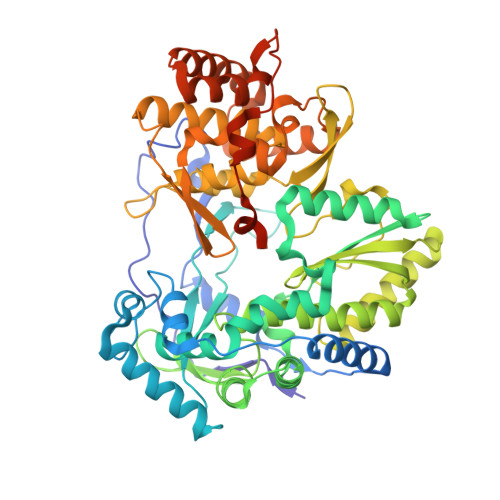
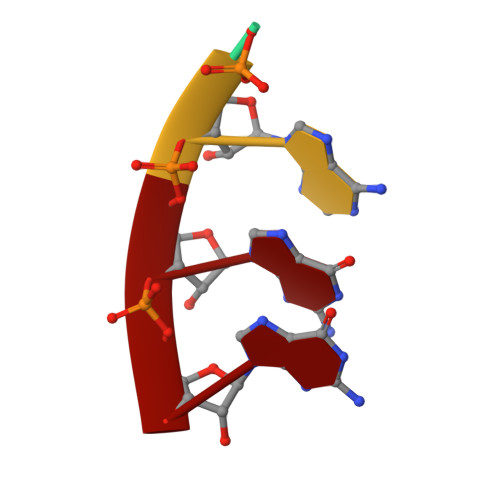
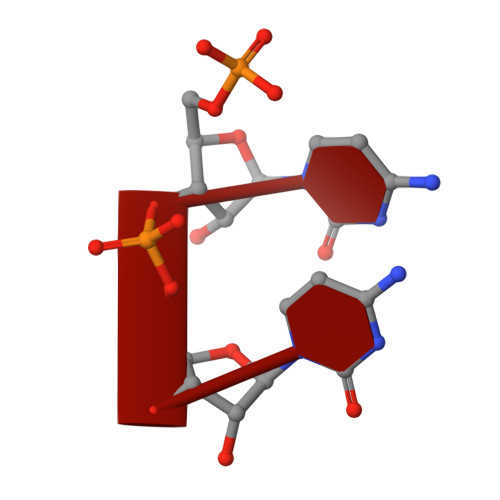
Structural basis for RNA replication by the hepatitis C virus polymerase.
Appleby, T.C., Perry, J.K., Murakami, E., Barauskas, O., Feng, J., Cho, A., Fox, D., Wetmore, D.R., McGrath, M.E., Ray, A.S., Sofia, M.J., Swaminathan, S., Edwards, T.E.(2015) Science 347: 771-775
- PubMed: 25678663
- DOI: https://doi.org/10.1126/science.1259210
- Primary Citation of Related Structures:
4WT9, 4WTA, 4WTC, 4WTD, 4WTE, 4WTF, 4WTG, 4WTI, 4WTJ, 4WTK, 4WTL, 4WTM - PubMed Abstract:
Nucleotide analog inhibitors have shown clinical success in the treatment of hepatitis C virus (HCV) infection, despite an incomplete mechanistic understanding of NS5B, the viral RNA-dependent RNA polymerase. Here we study the details of HCV RNA replication by determining crystal structures of stalled polymerase ternary complexes with enzymes, RNA templates, RNA primers, incoming nucleotides, and catalytic metal ions during both primed initiation and elongation of RNA synthesis. Our analysis revealed that highly conserved active-site residues in NS5B position the primer for in-line attack on the incoming nucleotide. A β loop and a C-terminal membrane-anchoring linker occlude the active-site cavity in the apo state, retract in the primed initiation assembly to enforce replication of the HCV genome from the 3' terminus, and vacate the active-site cavity during elongation. We investigated the incorporation of nucleotide analog inhibitors, including the clinically active metabolite formed by sofosbuvir, to elucidate key molecular interactions in the active site.
- Gilead Sciences, 333 Lakeside Drive, Foster City, CA 94404, USA. todd.appleby@gilead.com tedwards@be4.com.
Organizational Affiliation: