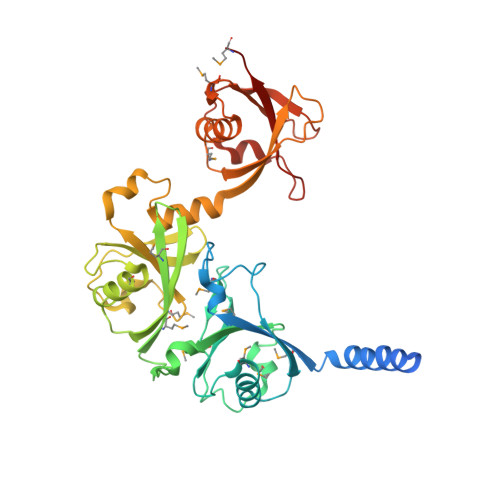
Crystal Structure of USP7 Ubiquitin-like Domains with an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target USP7.
Pfoh, R., Lacdao, I.K., Georges, A.A., Capar, A., Zheng, H., Frappier, L., Saridakis, V.(2015) PLoS Pathog 11: e1004950-e1004950
- PubMed: 26046769
- DOI: https://doi.org/10.1371/journal.ppat.1004950
- Primary Citation of Related Structures:
4WPH, 4WPI - PubMed Abstract:
Herpes simplex virus-1 immediate-early protein ICP0 activates viral genes during early stages of infection, affects cellular levels of multiple host proteins and is crucial for effective lytic infection. Being a RING-type E3 ligase prone to auto-ubiquitination, ICP0 relies on human deubiquitinating enzyme USP7 for protection against 26S proteasomal mediated degradation. USP7 is involved in apoptosis, epigenetics, cell proliferation and is targeted by several herpesviruses. Several USP7 partners, including ICP0, GMPS, and UHRF1, interact through its C-terminal domain (CTD), which contains five ubiquitin-like (Ubl) structures. Despite the fact that USP7 has emerged as a drug target for cancer therapy, structural details of USP7 regulation and the molecular mechanism of interaction at its CTD have remained elusive. Here, we mapped the binding site between an ICP0 peptide and USP7 and determined the crystal structure of the first three Ubl domains bound to the ICP0 peptide, which showed that ICP0 binds to a loop on Ubl2. Sequences similar to the USP7-binding site in ICP0 were identified in GMPS and UHRF1 and shown to bind USP7-CTD through Ubl2. In addition, co-immunoprecipitation assays in human cells comparing binding to USP7 with and without a Ubl2 mutation, confirmed the importance of the Ubl2 binding pocket for binding ICP0, GMPS and UHRF1. Therefore we have identified a novel mechanism of USP7 recognition that is used by both viral and cellular proteins. Our structural information was used to generate a model of near full-length USP7, showing the relative position of the ICP0/GMPS/UHRF1 binding pocket and the structural basis by which it could regulate enzymatic activity.
- Department of Biology, York University, Toronto, Ontario, Canada.
Organizational Affiliation: