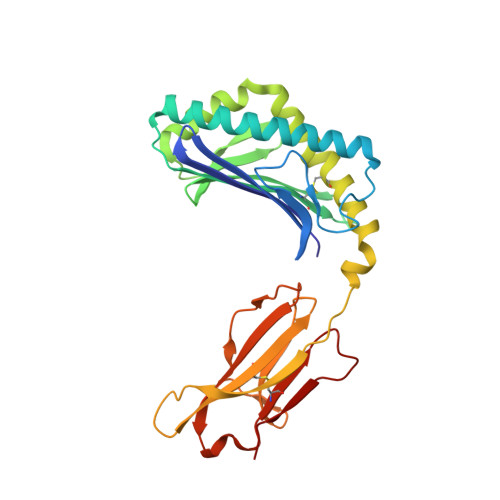
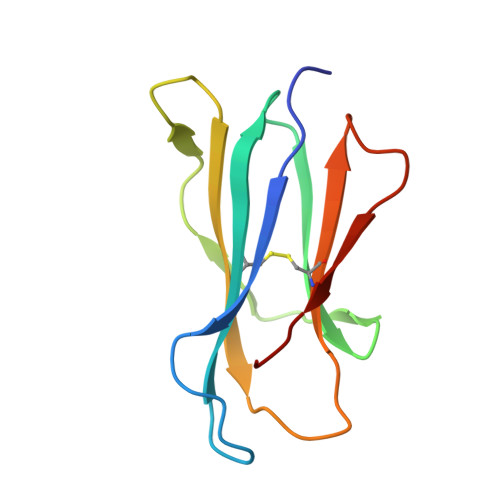
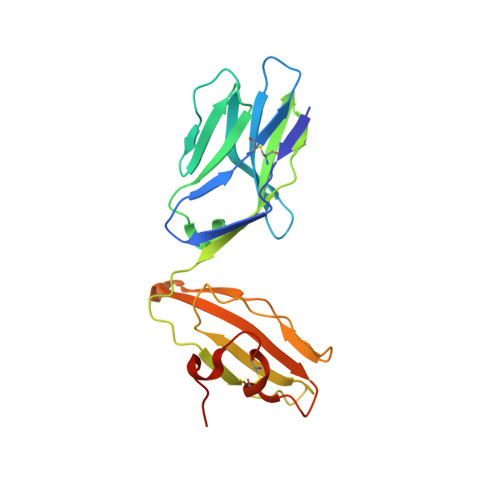
The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
Pellicci, D.G., Uldrich, A.P., Le Nours, J., Ross, F., Chabrol, E., Eckle, S.B., de Boer, R., Lim, R.T., McPherson, K., Besra, G., Howell, A.R., Moretta, L., McCluskey, J., Heemskerk, M.H., Gras, S., Rossjohn, J., Godfrey, D.I.(2014) J Exp Medicine 211: 2599-2615
- PubMed: 25452463
- DOI: https://doi.org/10.1084/jem.20141764
- Primary Citation of Related Structures:
4QRR, 4WNQ, 4WO4 - PubMed Abstract:
αβ and γδ T cells are disparate T cell lineages that can respond to distinct antigens (Ags) via the use of the αβ and γδ T cell Ag receptors (TCRs), respectively. Here we characterize a population of human T cells, which we term δ/αβ T cells, expressing TCRs comprised of a TCR-δ variable gene (Vδ1) fused to joining α and constant α domains, paired with an array of TCR-β chains. We demonstrate that these cells, which represent ∼50% of all Vδ1(+) human T cells, can recognize peptide- and lipid-based Ags presented by human leukocyte antigen (HLA) and CD1d, respectively. Similar to type I natural killer T (NKT) cells, CD1d-lipid Ag-reactive δ/αβ T cells recognized α-galactosylceramide (α-GalCer); however, their fine specificity for other lipid Ags presented by CD1d, such as α-glucosylceramide, was distinct from type I NKT cells. Thus, δ/αβTCRs contribute new patterns of Ag specificity to the human immune system. Furthermore, we provide the molecular bases of how δ/αβTCRs bind to their targets, with the Vδ1-encoded region providing a major contribution to δ/αβTCR binding. Our findings highlight how components from αβ and γδTCR gene loci can recombine to confer Ag specificity, thus expanding our understanding of T cell biology and TCR diversity.
- Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity and Australian Research Council Centre of Excellence in Advanced Molecular Imaging, University of Melbourne, Parkville, Victoria 3010, Australia Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity and Australian Research Council Centre of Excellence in Advanced Molecular Imaging, University of Melbourne, Parkville, Victoria 3010, Australia.
Organizational Affiliation: