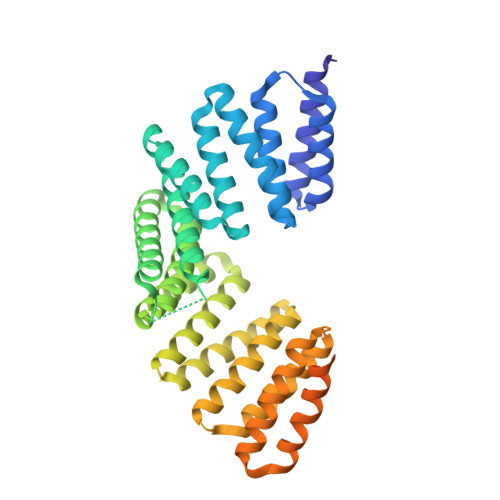
Structural basis for the recognition of the scaffold protein Frmpd4/Preso1 by the TPR domain of the adaptor protein LGN
Takayanagi, H., Yuzawa, S., Sumimoto, H.(2015) Acta Crystallogr F Struct Biol Commun 71: 175-183
- PubMed: 25664792
- DOI: https://doi.org/10.1107/S2053230X14028143
- Primary Citation of Related Structures:
4WND, 4WNE, 4WNF, 4WNG - PubMed Abstract:
The adaptor protein LGN interacts via the N-terminal domain comprising eight tetratricopeptide-repeat (TPR) motifs with its partner proteins mInsc, NuMA, Frmpd1 and Frmpd4 in a mutually exclusive manner. Here, the crystal structure of the LGN TPR domain in complex with human Frmpd4 is described at 1.5 Å resolution. In the complex, the LGN-binding region of Frmpd4 (amino-acid residues 990-1011) adopts an extended structure that runs antiparallel to LGN along the concave surface of the superhelix formed by the TPR motifs. Comparison with the previously determined structures of the LGN-Frmpd1, LGN-mInsc and LGN-NuMA complexes reveals that these partner proteins interact with LGN TPR1-6 via a common core binding region with consensus sequence (E/Q)XEX4-5(E/D/Q)X1-2(K/R)X0-1(V/I). In contrast to Frmpd1, Frmpd4 makes additional contacts with LGN via regions N- and C-terminal to the core sequence. The N-terminal extension is replaced by a specific α-helix in mInsc, which drastically increases the direct contacts with LGN TPR7/8, consistent with the higher affinity of mInsc for LGN. A crystal structure of Frmpd4-bound LGN in an oxidized form is also reported, although oxidation does not appear to strongly affect the interaction with Frmpd4.
- Department of Biochemistry, Kyushu University Graduate School of Medical Sciences, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Organizational Affiliation: