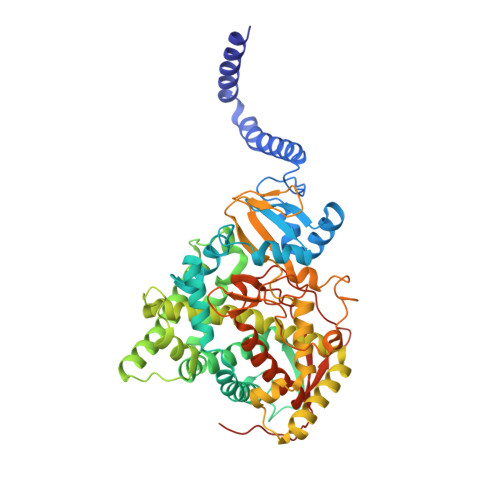
Structural Insights into Binding of the Antifungal Drug Fluconazole to Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Sagatova, A.A., Keniya, M.V., Wilson, R.K., Monk, B.C., Tyndall, J.D.(2015) Antimicrob Agents Chemother 59: 4982-4989
- PubMed: 26055382
- DOI: https://doi.org/10.1128/AAC.00925-15
- Primary Citation of Related Structures:
4WMZ - PubMed Abstract:
Infections by fungal pathogens such as Candida albicans and Aspergillus fumigatus and their resistance to triazole drugs are major concerns. Fungal lanosterol 14α-demethylase belongs to the CYP51 class in the cytochrome P450 superfamily of enzymes. This monospanning bitopic membrane protein is involved in ergosterol biosynthesis and is the primary target of azole antifungal drugs, including fluconazole. The lack of high-resolution structural information for this drug target from fungal pathogens has been a limiting factor for the design of modified triazole drugs that will overcome resistance. Here we report the X-ray structure of full-length Saccharomyces cerevisiae lanosterol 14α-demethylase in complex with fluconazole at a resolution of 2.05 Å. This structure shows the key interactions involved in fluconazole binding and provides insight into resistance mechanisms by revealing a water-mediated hydrogen bonding network between the drug and tyrosine 140, a residue frequently found mutated to histidine or phenylalanine in resistant clinical isolates.
- Sir John Walsh Research Institute, University of Otago, Dunedin, New Zealand.
Organizational Affiliation: