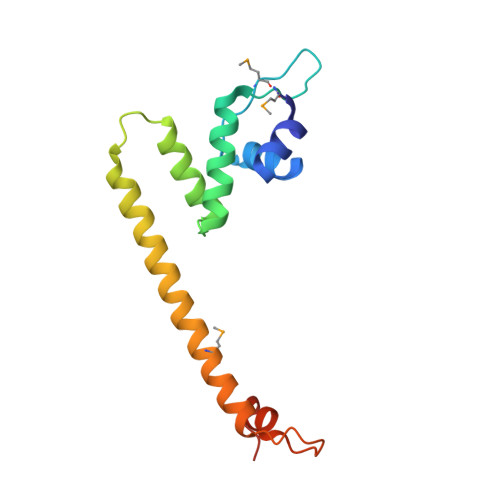
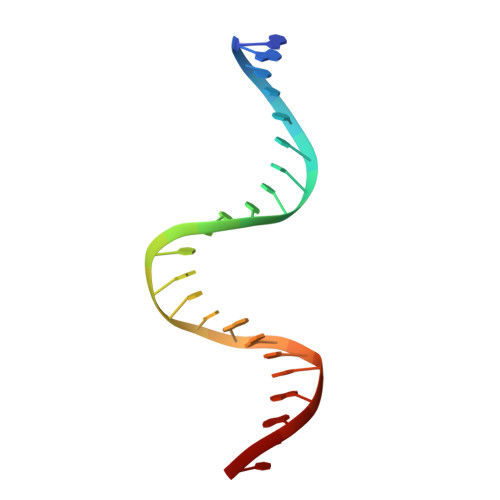
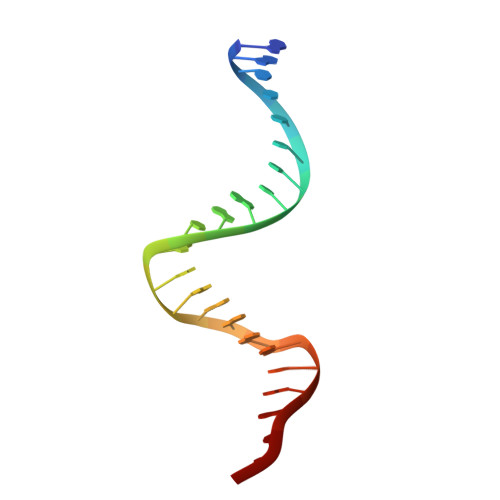
TRANSCRIPTION. Allosteric transcriptional regulation via changes in the overall topology of the core promoter.
Philips, S.J., Canalizo-Hernandez, M., Yildirim, I., Schatz, G.C., Mondragon, A., O'Halloran, T.V.(2015) Science 349: 877-881
- PubMed: 26293965
- DOI: https://doi.org/10.1126/science.aaa9809
- Primary Citation of Related Structures:
4WLS, 4WLW - PubMed Abstract:
Many transcriptional activators act at a distance from core promoter elements and work by recruiting RNA polymerase through protein-protein interactions. We show here how the prokaryotic regulatory protein CueR both represses and activates transcription by differentially modulating local DNA structure within the promoter. Structural studies reveal that the repressor state slightly bends the promoter DNA, precluding optimal RNA polymerase-promoter recognition. Upon binding a metal ion in the allosteric site, CueR switches into an activator conformation. It maintains all protein-DNA contacts but introduces torsional stresses that kink and undertwist the promoter, stabilizing an A-form DNA-like conformation. These factors switch on and off transcription by exerting dynamic control of DNA stereochemistry, reshaping the core promoter and making it a better or worse substrate for polymerase.
- Department of Molecular Biosciences, Northwestern University, Evanston, IL 60208, USA.
Organizational Affiliation: