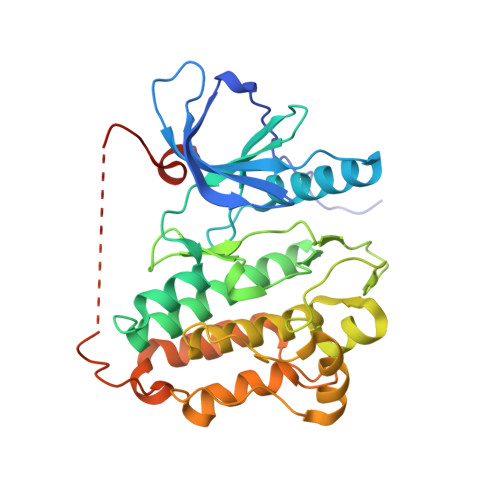
Crystal structure of EGFR 696-1022 T790M in complex with QL-X138
Wu, H., Hu, C., Wang, A., Weisberg, E.E., Chen, Y., Yun, C.H., Wang, W., Liu, X., Tian, B., Wang, J., Zhao, Z., Liang, Y., Li, B., Wang, L., Wang, B., Chen, C., Buhrlage, S.J., Fernadners, S.M., Griffin, J., Brown, J.R., Eck, M.J., Liu, J., Liu, Q., Gray, N.S.To be published.