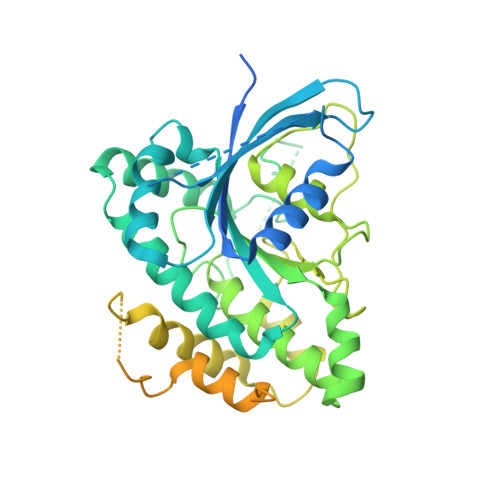
A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region.
Schroeder, C.M., Ostrem, J.M., Hertz, N.T., Vale, R.D.(2014) Elife 3: e03351-e03351
- PubMed: 25272277
- DOI: https://doi.org/10.7554/eLife.03351
- Primary Citation of Related Structures:
4W7G - PubMed Abstract:
Cytoplasmic dynein, a microtubule-based motor protein, transports many intracellular cargos by means of its light intermediate chain (LIC). In this study, we have determined the crystal structure of the conserved LIC domain, which binds the motor heavy chain, from a thermophilic fungus. We show that the LIC has a Ras-like fold with insertions that distinguish it from Ras and other previously described G proteins. Despite having a G protein fold, the fungal LIC has lost its ability to bind nucleotide, while the human LIC1 binds GDP preferentially over GTP. We show that the LIC G domain binds the dynein heavy chain using a conserved patch of aromatic residues, whereas the less conserved C-terminal domain binds several Rab effectors involved in membrane transport. These studies provide the first structural information and insight into the evolutionary origin of the LIC as well as revealing how this critical subunit connects the dynein motor to cargo.
- Department of Cellular and Molecular Pharmacology, Howard Hughes Medical Institute, University of California, San Francisco, San Francisco, United States.
Organizational Affiliation: