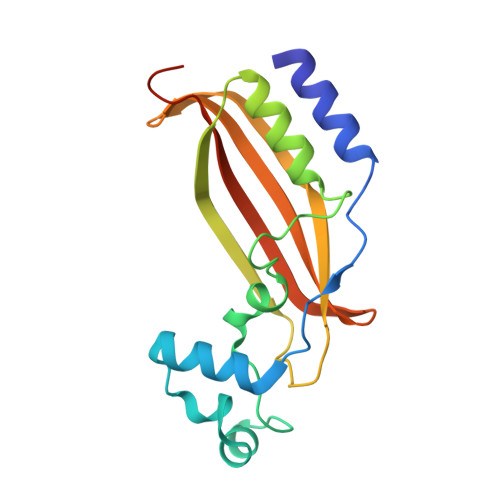
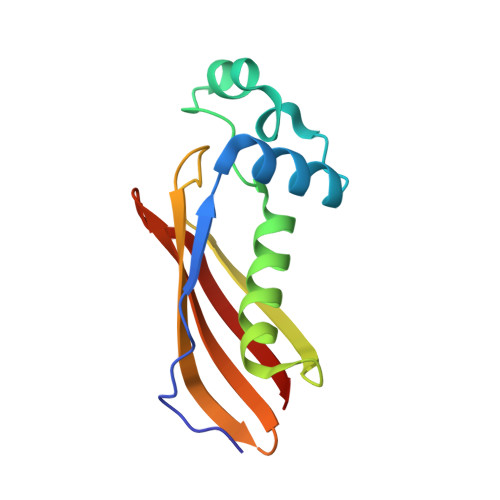
A Distinct MaoC-like Enoyl-CoA Hydratase Architecture Mediates Cholesterol Catabolism in Mycobacterium tuberculosis.
Yang, M., Guja, K.E., Thomas, S.T., Garcia-Diaz, M., Sampson, N.S.(2014) ACS Chem Biol 9: 2632-2645
- PubMed: 25203216
- DOI: https://doi.org/10.1021/cb500232h
- Primary Citation of Related Structures:
4W78, 4WNB - PubMed Abstract:
The Mycobacterium tuberculosis (Mtb) igr operon plays an essential role in Mtb cholesterol metabolism, which is critical for pathogenesis during the latent stage of Mtb infection. Here we report the first structure of a heterotetrameric MaoC-like enoyl-CoA hydratase, ChsH1-ChsH2, which is encoded by two adjacent genes from the igr operon. We demonstrate that ChsH1-ChsH2 catalyzes the hydration of a steroid enoyl-CoA, 3-oxo-4,17-pregnadiene-20-carboxyl-CoA, in the modified β-oxidation pathway for cholesterol side chain degradation. The ligand-bound and apoenzyme structures of ChsH1-ChsH2(N) reveal an unusual, modified hot-dog fold with a severely truncated central α-helix that creates an expanded binding site to accommodate the bulkier steroid ring system. The structures show quaternary structure shifts that accommodate the four rings of the steroid substrate and offer an explanation for why the unusual heterotetrameric assembly is utilized for hydration of this steroid. The unique αβ heterodimer architecture utilized by ChsH1-ChsH2 to bind its distinctive substrate highlights an opportunity for the development of new antimycobacterial drugs that target a pathway specific to Mtb.
- Department of Chemistry, Stony Brook University , Stony Brook, New York 11794-3400, United States.
Organizational Affiliation: