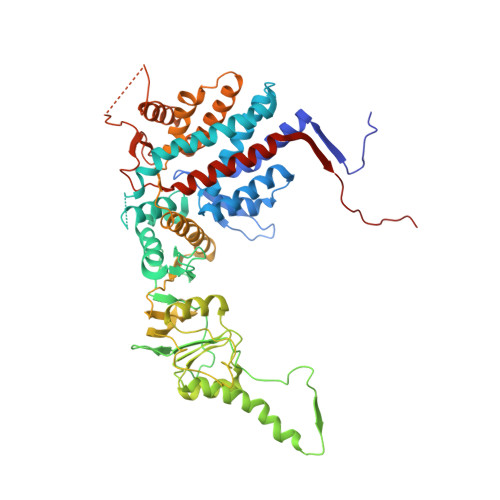
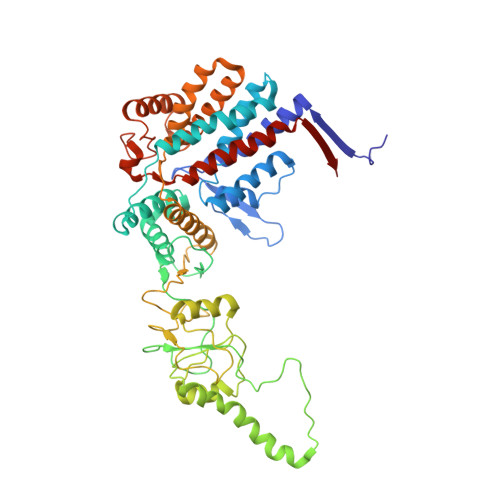
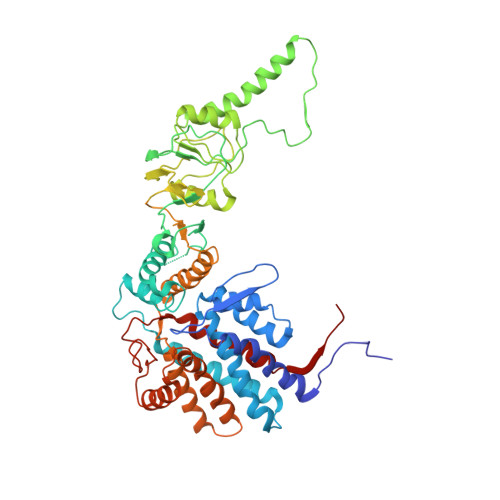
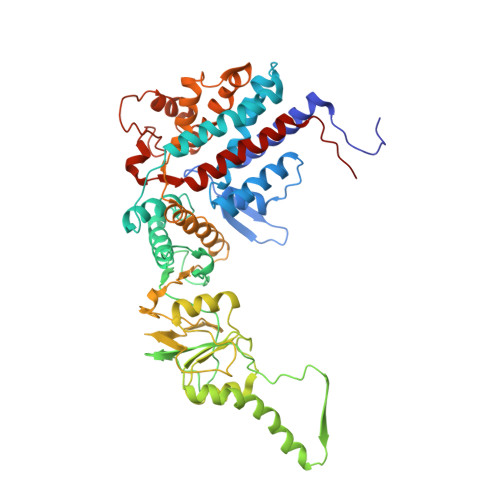
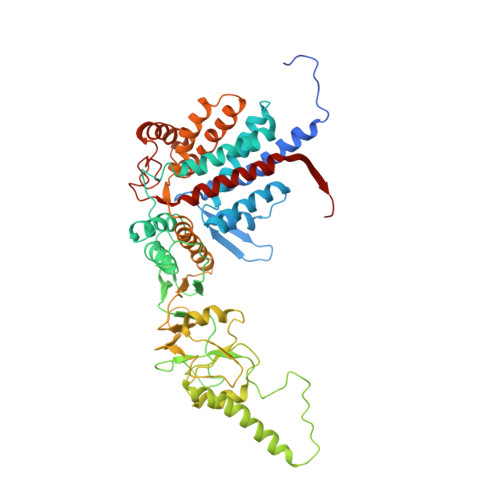
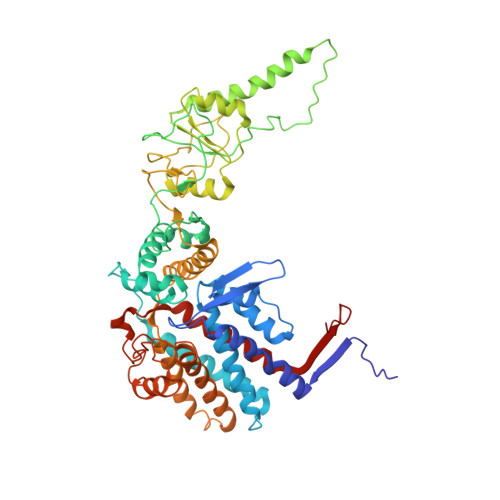
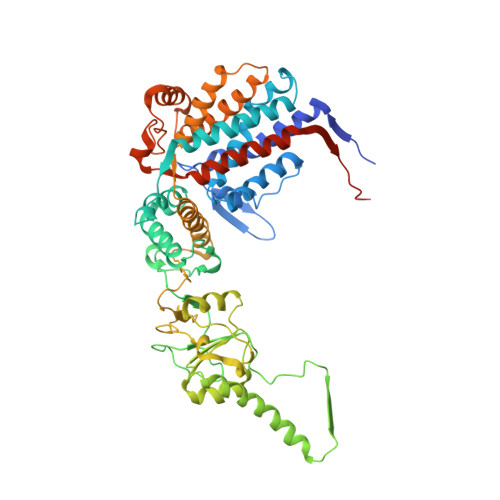
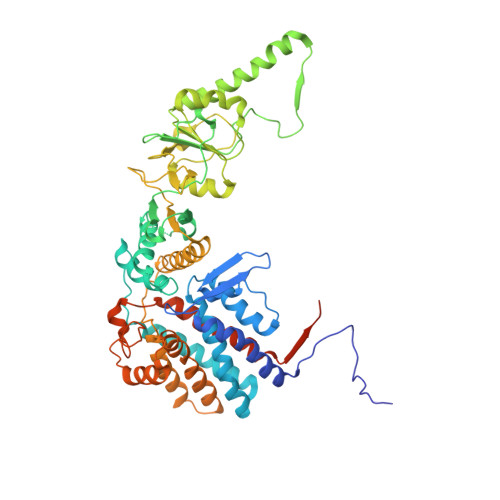
The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins.
Dekker, C., Roe, S.M., McCormack, E.A., Beuron, F., Pearl, L.H., Willison, K.R.(2011) EMBO J 30: 3078-3090
- PubMed: 21701561
- DOI: https://doi.org/10.1038/emboj.2011.208
- Primary Citation of Related Structures:
4V81 - PubMed Abstract:
The cytosolic chaperonin CCT is a 1-MDa protein-folding machine essential for eukaryotic life. The CCT interactome shows involvement in folding and assembly of a small range of proteins linked to essential cellular processes such as cytoskeleton assembly and cell-cycle regulation. CCT has a classic chaperonin architecture, with two heterogeneous 8-membered rings stacked back-to-back, enclosing a folding cavity. However, the mechanism by which CCT assists folding is distinct from other chaperonins, with no hydrophobic wall lining a potential Anfinsen cage, and a sequential rather than concerted ATP hydrolysis mechanism. We have solved the crystal structure of yeast CCT in complex with actin at 3.8 Å resolution, revealing the subunit organisation and the location of discrete patches of co-evolving 'signature residues' that mediate specific interactions between CCT and its substrates. The intrinsic asymmetry is revealed by the structural individuality of the CCT subunits, which display unique configurations, substrate binding properties, ATP-binding heterogeneity and subunit-subunit interactions. The location of the evolutionarily conserved N-terminus of Cct5 on the outside of the barrel, confirmed by mutational studies, is unique to eukaryotic cytosolic chaperonins.
- Section of Cell and Molecular Biology, Chester Beatty Laboratories, Institute of Cancer Research, London, UK.
Organizational Affiliation: