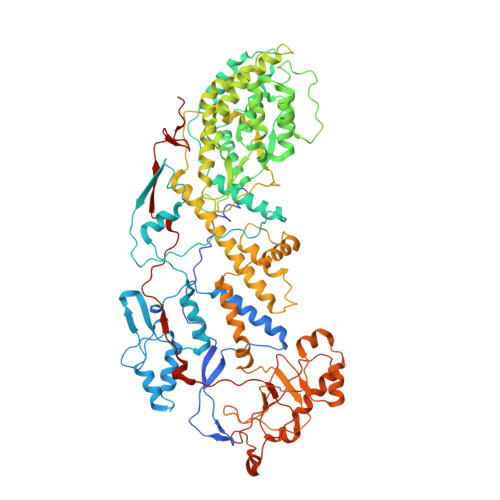
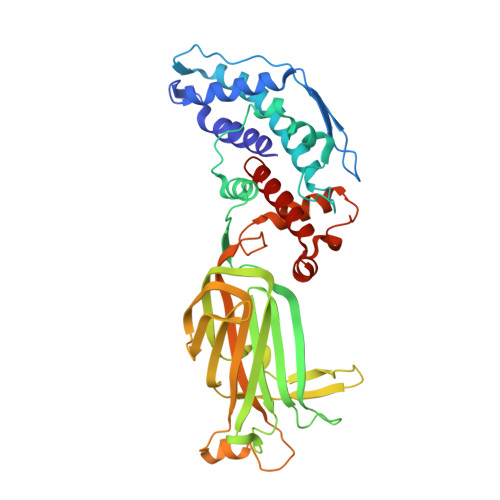
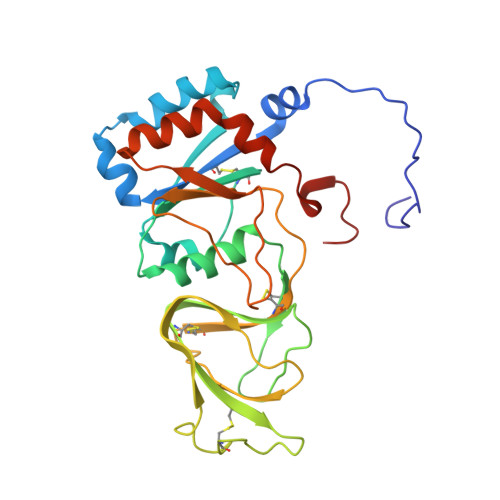
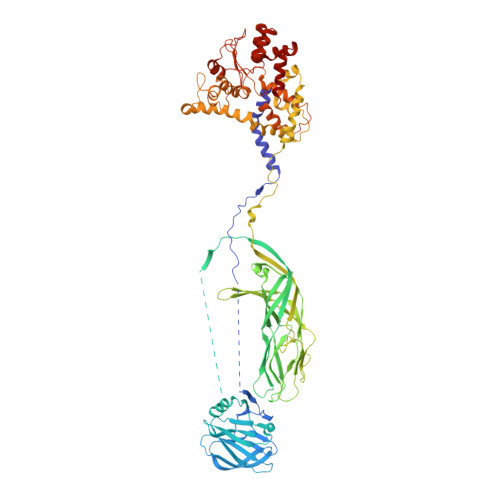
Atomic model of an infectious rotavirus particle.
Settembre, E.C., Chen, J.Z., Dormitzer, P.R., Grigorieff, N., Harrison, S.C.(2011) EMBO J 30: 408-416
- PubMed: 21157433
- DOI: https://doi.org/10.1038/emboj.2010.322
- Primary Citation of Related Structures:
4V7Q - PubMed Abstract:
Non-enveloped viruses of different types have evolved distinct mechanisms for penetrating a cellular membrane during infection. Rotavirus penetration appears to occur by a process resembling enveloped-virus fusion: membrane distortion linked to conformational changes in a viral protein. Evidence for such a mechanism comes from crystallographic analyses of fragments of VP4, the rotavirus-penetration protein, and infectivity analyses of structure-based VP4 mutants. We describe here the structure of an infectious rotavirus particle determined by electron cryomicroscopy (cryoEM) and single-particle analysis at about 4.3 Å resolution. The cryoEM image reconstruction permits a nearly complete trace of the VP4 polypeptide chain, including the positions of most side chains. It shows how the two subfragments of VP4 (VP8(*) and VP5(*)) retain their association after proteolytic cleavage, reveals multiple structural roles for the β-barrel domain of VP5(*), and specifies interactions of VP4 with other capsid proteins. The virion model allows us to integrate structural and functional information into a coherent mechanism for rotavirus entry.
- Laboratory of Molecular Medicine, Children's Hospital, Boston, MA, USA.
Organizational Affiliation: