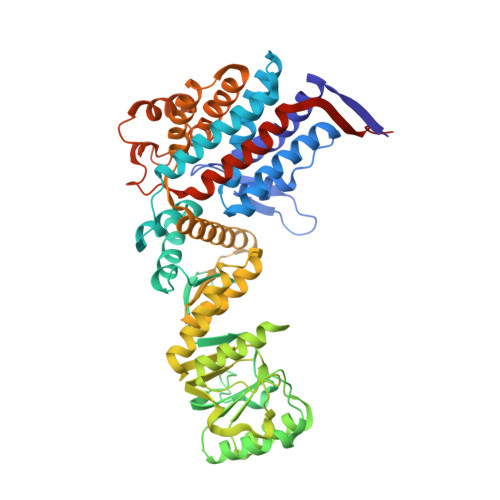
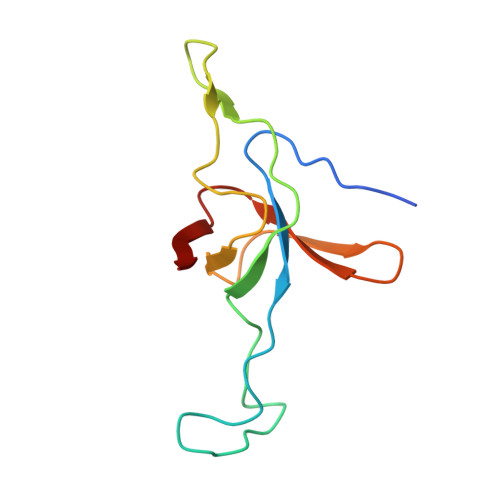
Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity
Shimamura, T., Koike-Takeshita, A., Yokoyama, K., Masui, R., Murai, N., Yoshida, M., Taguchi, H., Iwata, S.(2004) Structure 12: 1471-1480
- PubMed: 15296740
- DOI: https://doi.org/10.1016/j.str.2004.05.020
- Primary Citation of Related Structures:
4V4O - PubMed Abstract:
The chaperonins GroEL and GroES are essential mediators of protein folding. GroEL binds nonnative protein, ATP, and GroES, generating a ternary complex in which protein folding occurs within the cavity capped by GroES (cis-cavity). We determined the crystal structure of the native GroEL-GroES-ADP homolog from Thermus thermophilus, with substrate proteins in the cis-cavity, at 2.8 A resolution. Twenty-four in vivo substrate proteins within the cis-cavity were identified from the crystals. The structure around the cis-cavity, which encapsulates substrate proteins, shows significant differences from that observed for the substrate-free Escherichia coli GroEL-GroES complex. The apical domain around the cis-cavity of the Thermus GroEL-GroES complex exhibits a large deviation from the 7-fold symmetry. As a result, the GroEL-GroES interface differs considerably from the previously reported E. coli GroEL-GroES complex, including a previously unknown contact between GroEL and GroES.
- Department of Biological Sciences, Imperial College London, London SW7 2AZ, United Kingdom.
Organizational Affiliation: