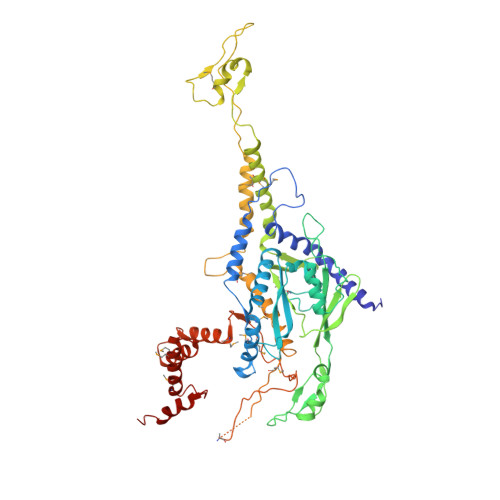
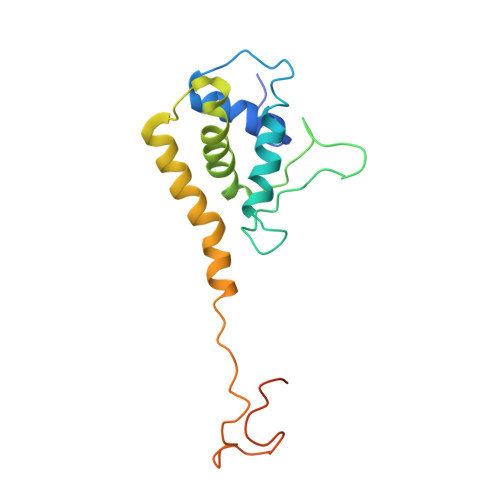
Three-dimensional structure of a viral genome-delivery portal vertex.
Olia, A.S., Prevelige, P.E., Johnson, J.E., Cingolani, G.(2011) Nat Struct Mol Biol 18: 597-603
- PubMed: 21499245
- DOI: https://doi.org/10.1038/nsmb.2023
- Primary Citation of Related Structures:
3LJ5, 4V4K - PubMed Abstract:
DNA viruses such as bacteriophages and herpesviruses deliver their genome into and out of the capsid through large proteinaceous assemblies, known as portal proteins. Here, we report two snapshots of the dodecameric portal protein of bacteriophage P22. The 3.25-Å-resolution structure of the portal-protein core bound to 12 copies of gene product 4 (gp4) reveals a ~1.1-MDa assembly formed by 24 proteins. Unexpectedly, a lower-resolution structure of the full-length portal protein unveils the unique topology of the C-terminal domain, which forms a ~200-Å-long α-helical barrel. This domain inserts deeply into the virion and is highly conserved in the Podoviridae family. We propose that the barrel domain facilitates genome spooling onto the interior surface of the capsid during genome packaging and, in analogy to a rifle barrel, increases the accuracy of genome ejection into the host cell.
- Department of Biological Sciences, Purdue University, West Lafayette, Indiana, USA.
Organizational Affiliation: