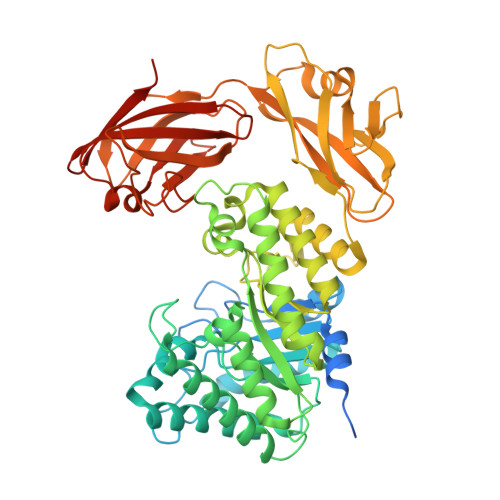
Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
Venditto, I., Najmudin, S., Luis, A.S., Ferreira, L.M., Sakka, K., Knox, J.P., Gilbert, H.J., Fontes, C.M.(2015) J Biological Chem 290: 10572
- PubMed: 25713075
- DOI: https://doi.org/10.1074/jbc.M115.637827
- Primary Citation of Related Structures:
4UZ8, 4UZN, 4V2X - PubMed Abstract:
Structural carbohydrates comprise an extraordinary source of energy that remains poorly utilized by the biofuel sector as enzymes have restricted access to their substrates within the intricacy of plant cell walls. Carbohydrate active enzymes (CAZYmes) that target recalcitrant polysaccharides are modular enzymes containing noncatalytic carbohydrate-binding modules (CBMs) that direct enzymes to their cognate substrate, thus potentiating catalysis. In general, CBMs are functionally and structurally autonomous from their associated catalytic domains from which they are separated through flexible linker sequences. Here, we show that a C-terminal CBM46 derived from BhCel5B, a Bacillus halodurans endoglucanase, does not interact with β-glucans independently but, uniquely, acts cooperatively with the catalytic domain of the enzyme in substrate recognition. The structure of BhCBM46 revealed a β-sandwich fold that abuts onto the region of the substrate binding cleft upstream of the active site. BhCBM46 as a discrete entity is unable to bind to β-glucans. Removal of BhCBM46 from BhCel5B, however, abrogates binding to β-1,3-1,4-glucans while substantially decreasing the affinity for decorated β-1,4-glucan homopolymers such as xyloglucan. The CBM46 was shown to contribute to xyloglucan hydrolysis only in the context of intact plant cell walls, but it potentiates enzymatic activity against purified β-1,3-1,4-glucans in solution or within the cell wall. This report reveals the mechanism by which a CBM can promote enzyme activity through direct interaction with the substrate or by targeting regions of the plant cell wall where the target glucan is abundant.
- From the Centro Interdisciplinar de Investigação em Sanidade Animal, Faculdade de Medicina Veterinária, ULisboa, Pólo Universitário do Alto da Ajuda, Avenida da Universidade Técnica, 1300-477 Lisboa, Portugal.
Organizational Affiliation: