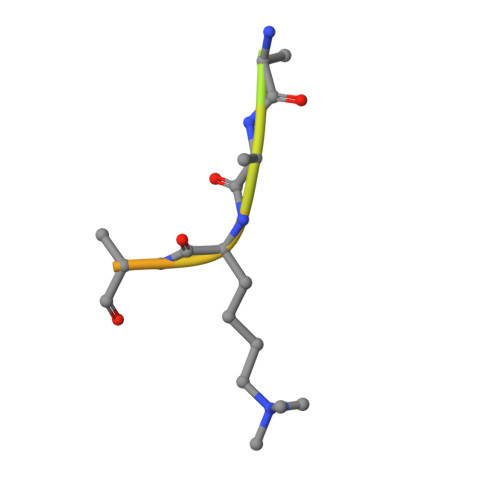
Studies on the Catalytic Domains of Multiple Jmjc Oxygenases Using Peptide Substrates.
Williams, S.T., Walport, L.J., Hopkinson, R.J., Madden, S.K., Chowdhury, R., Schofield, C.J., Kawamura, A.(2014) Epigenetics 9: 1596
- PubMed: 25625844
- DOI: https://doi.org/10.4161/15592294.2014.983381
- Primary Citation of Related Structures:
4V2V, 4V2W - PubMed Abstract:
The JmjC-domain-containing 2-oxoglutarate-dependent oxygenases catalyze protein hydroxylation and N(ϵ)-methyllysine demethylation via hydroxylation. A subgroup of this family, the JmjC lysine demethylases (JmjC KDMs) are involved in histone modifications at multiple sites. There are conflicting reports as to the substrate selectivity of some JmjC oxygenases with respect to KDM activities. In this study, a panel of modified histone H3 peptides was tested for demethylation against 15 human JmjC-domain-containing proteins. The results largely confirmed known N(ϵ)-methyllysine substrates. However, the purified KDM4 catalytic domains showed greater substrate promiscuity than previously reported (i.e., KDM4A was observed to catalyze demethylation at H3K27 as well as H3K9/K36). Crystallographic analyses revealed that the N(ϵ)-methyllysine of an H3K27me3 peptide binds similarly to N(ϵ)-methyllysines of H3K9me3/H3K36me3 with KDM4A. A subgroup of JmjC proteins known to catalyze hydroxylation did not display demethylation activity. Overall, the results reveal that the catalytic domains of the KDM4 enzymes may be less selective than previously identified. They also draw a distinction between the N(ϵ)-methyllysine demethylation and hydroxylation activities within the JmjC subfamily. These results will be of use to those working on functional studies of the JmjC enzymes.
- a Chemistry Research Laboratory ; Oxford , UK.
Organizational Affiliation: