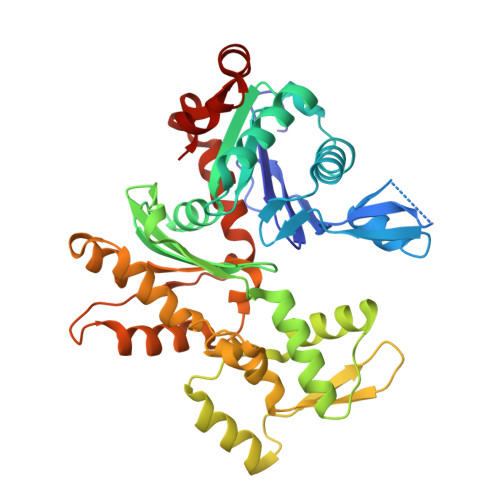
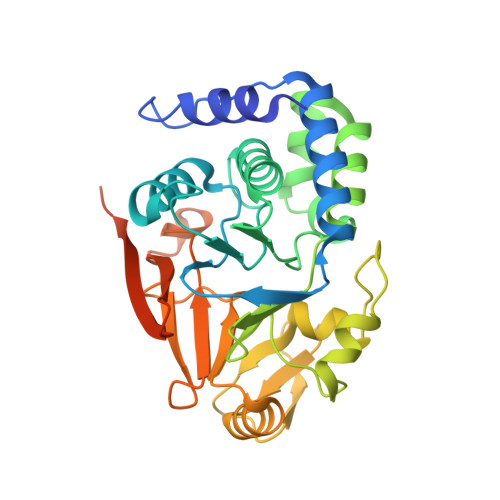
G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Chen, R., Rato, C., Yan, Y., Crespillo-Casado, A., Clarke, H.J., Harding, H.P., Marciniak, S.J., Read, R.J., Ron, D.(2015) Elife 4
- PubMed: 25774600
- DOI: https://doi.org/10.7554/eLife.04871
- Primary Citation of Related Structures:
4V0U, 4V0V, 4V0W, 4V0X - PubMed Abstract:
Dephosphorylation of eukaryotic translation initiation factor 2a (eIF2a) restores protein synthesis at the waning of stress responses and requires a PP1 catalytic subunit and a regulatory subunit, PPP1R15A/GADD34 or PPP1R15B/CReP. Surprisingly, PPP1R15-PP1 binary complexes reconstituted in vitro lacked substrate selectivity. However, selectivity was restored by crude cell lysate or purified G-actin, which joined PPP1R15-PP1 to form a stable ternary complex. In crystal structures of the non-selective PPP1R15B-PP1G complex, the functional core of PPP1R15 made multiple surface contacts with PP1G, but at a distance from the active site, whereas in the substrate-selective ternary complex, actin contributes to one face of a platform encompassing the active site. Computational docking of the N-terminal lobe of eIF2a at this platform placed phosphorylated serine 51 near the active site. Mutagenesis of predicted surface-contacting residues enfeebled dephosphorylation, suggesting that avidity for the substrate plays an important role in imparting specificity on the PPP1R15B-PP1G-actin ternary complex.
- Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom.
Organizational Affiliation: