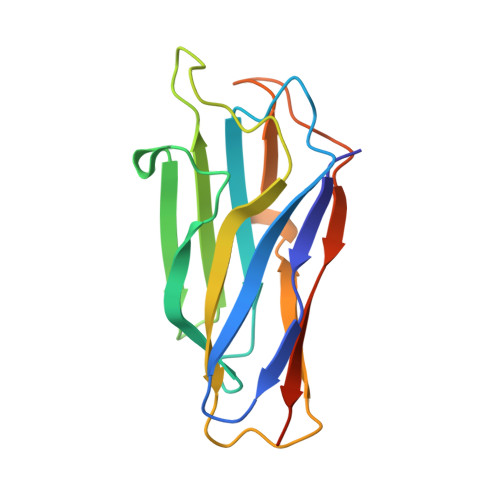
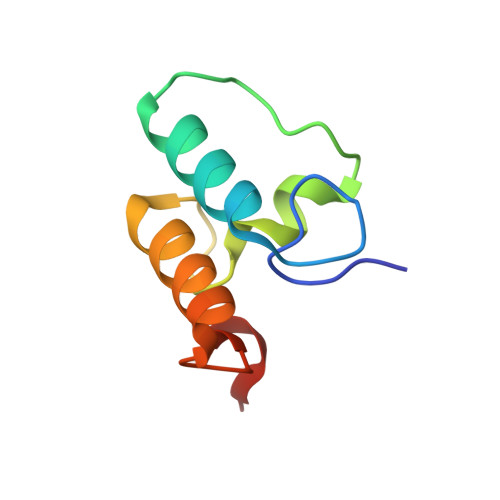
Cell-surface Attachment of Bacterial Multienzyme Complexes Involves Highly Dynamic Protein-Protein Anchors.
Cameron, K., Najmudin, S., Alves, V.D., Bayer, E.A., Smith, S.P., Bule, P., Waller, H., Ferreira, L.M., Gilbert, H.J., Fontes, C.M.(2015) J Biological Chem 290: 13578-13590
- PubMed: 25855788
- DOI: https://doi.org/10.1074/jbc.M114.633339
- Primary Citation of Related Structures:
4UYP, 4UYQ - PubMed Abstract:
Protein-protein interactions play a pivotal role in the assembly of the cellulosome, one of nature's most intricate nanomachines dedicated to the depolymerization of complex carbohydrates. The integration of cellulosomal components usually occurs through the binding of type I dockerin modules located at the C terminus of the enzymes to cohesin modules located in the primary scaffoldin subunit. Cellulosomes are typically recruited to the cell surface via type II cohesin-dockerin interactions established between primary and cell-surface anchoring scaffoldin subunits. In contrast with type II interactions, type I dockerins usually display a dual binding mode that may allow increased conformational flexibility during cellulosome assembly. Acetivibrio cellulolyticus produces a highly complex cellulosome comprising an unusual adaptor scaffoldin, ScaB, which mediates the interaction between the primary scaffoldin, ScaA, through type II cohesin-dockerin interactions and the anchoring scaffoldin, ScaC, via type I cohesin-dockerin interactions. Here, we report the crystal structure of the type I ScaB dockerin in complex with a type I ScaC cohesin in two distinct orientations. The data show that the ScaB dockerin displays structural symmetry, reflected by the presence of two essentially identical binding surfaces. The complex interface is more extensive than those observed in other type I complexes, which results in an ultra-high affinity interaction (Ka ∼10(12) M). A subset of ScaB dockerin residues was also identified as modulating the specificity of type I cohesin-dockerin interactions in A. cellulolyticus. This report reveals that recruitment of cellulosomes onto the cell surface may involve dockerins presenting a dual binding mode to incorporate additional flexibility into the quaternary structure of highly populated multienzyme complexes.
- From the CIISA-Faculdade de Medicina Veterinária, ULisboa, Pólo Universitário do Alto da Ajuda, Avenida da Universidade Técnica, 1300-477 Lisboa, Portugal.
Organizational Affiliation: