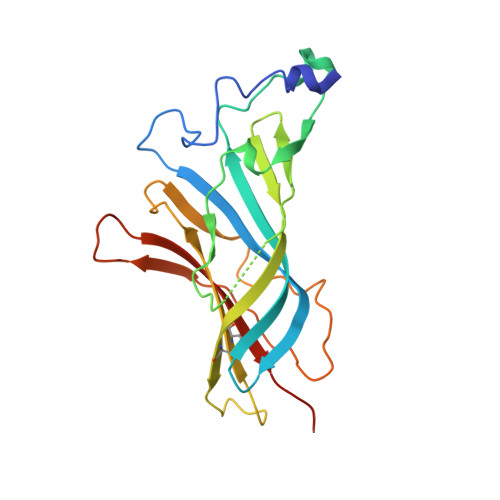
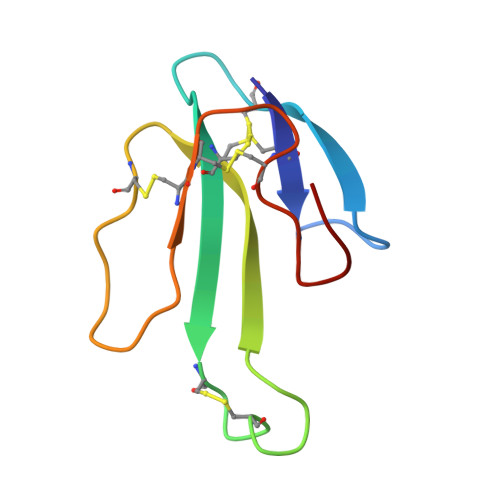
Crystal Structures of Free and Antagonist-Bound States of Human Alpha9 Nicotinic Receptor Extracellular Domain
Zouridakis, M., Giastas, P., Zarkadas, E., Chroni-Tzartou, D., Bregestovski, P., Tzartos, S.J.(2014) Nat Struct Mol Biol 21: 976
- PubMed: 25282151
- DOI: https://doi.org/10.1038/nsmb.2900
- Primary Citation of Related Structures:
4D01, 4UXU, 4UY2 - PubMed Abstract:
We determined the X-ray crystal structures of the extracellular domain (ECD) of the monomeric state of human neuronal α9 nicotinic acetylcholine receptor (nAChR) and of its complexes with the antagonists methyllycaconitine and α-bungarotoxin at resolutions of 1.8 Å, 1.7 Å and 2.7 Å, respectively. The structure of the monomeric α9 ECD superimposed well with the structures of homologous proteins in pentameric assemblies, denoting native folding, despite the absence of a complementary subunit and transmembrane domain. The interaction motifs of both antagonists were similar to those in the complexes with homologous pentameric proteins, thus highlighting the major contribution of the principal side of α9 ECD to their binding. The structures revealed a functionally important β7-β10 strand interaction in α9-containing nAChRs, involving their unique Thr147, a hydration pocket similar to that of mouse α1 ECD and a membrane-facing network coordinated by the invariant Arg210.
- Department of Neurobiology, Hellenic Pasteur Institute, Athens, Greece.
Organizational Affiliation: