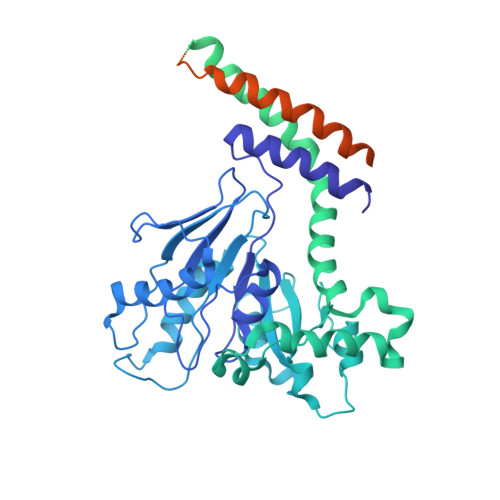
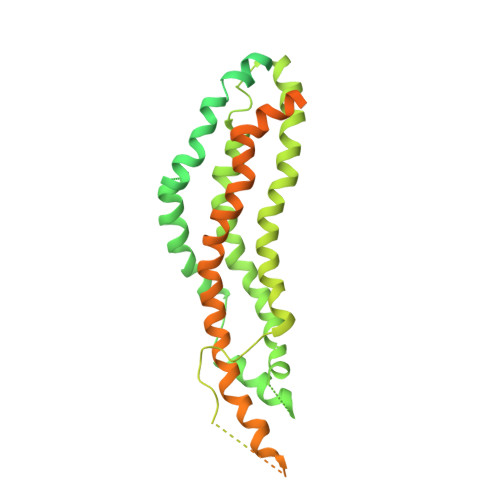
A Dynamin Mutant Defines a Superconstricted Prefission State.
Sundborger, A.C., Fang, S., Heymann, J.A., Ray, P., Chappie, J.S., Hinshaw, J.E.(2014) Cell Rep 8: 734
- PubMed: 25088425
- DOI: https://doi.org/10.1016/j.celrep.2014.06.054
- Primary Citation of Related Structures:
4UUD, 4UUK - PubMed Abstract:
Dynamin is a 100 kDa GTPase that organizes into helical assemblies at the base of nascent clathrin-coated vesicles. Formation of these oligomers stimulates the intrinsic GTPase activity of dynamin, which is necessary for efficient membrane fission during endocytosis. Recent evidence suggests that the transition state of dynamin's GTP hydrolysis reaction serves as a key determinant of productive fission. Here, we present the structure of a transition-state-defective dynamin mutant K44A trapped in a prefission state at 12.5 Å resolution. This structure constricts to 3.7 nm, reaching the theoretical limit required for spontaneous membrane fission. Computational docking indicates that the ground-state conformation of the dynamin polymer is sufficient to achieve this superconstricted prefission state and reveals how a two-start helical symmetry promotes the most efficient packing of dynamin tetramers around the membrane neck. These data suggest a model for the assembly and regulation of the minimal dynamin fission machine.
- Laboratory of Cell and Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: