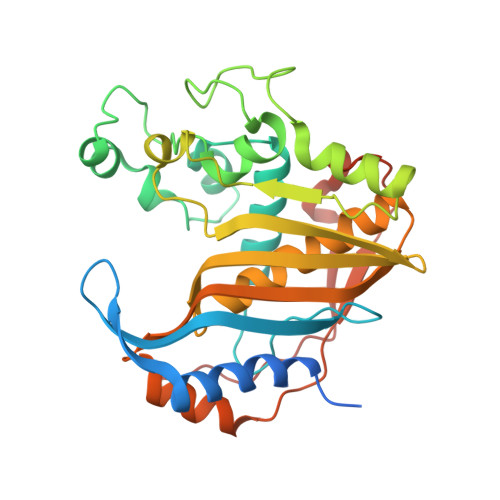
Crystal structure of the active form of native human thymidylate synthase in the absence of bound substrates.
Deschamps, P., Rety, S., Bareille, J., Leulliot, N.(2017) Acta Crystallogr F Struct Biol Commun 73: 336-341
- PubMed: 28580921
- DOI: https://doi.org/10.1107/S2053230X17007233
- Primary Citation of Related Structures:
4UP1 - PubMed Abstract:
Human thymidylate synthase (hTS) provides the sole de novo intracellular source of thymidine 5'-monophosphate (dTMP). hTS is required for DNA replication prior to cell division, making it an attractive target for anticancer chemotherapy and drug discovery. hTS binds 2'-deoxyuridine 5'-monophosphate (dUMP) and the folate co-substrate N 5 ,N 10 -methylenetetrahydrofolate (meTHF) in a pocket near the catalytic residue Cys195. The catalytic loop, which is composed of amino-acid residues 181-197, can adopt two distinct conformations related by a 180° rotation. In the active conformation Cys195 is close to the active site, while in the inactive conformation it is rotated and Cys195 is too distant from the active site for catalysis. Several hTS structures, either native or engineered, have been solved in the active conformation in complex with ligands or inhibitors and at different salt concentrations. However, apo hTS structures have been solved in an inactive conformation in high-salt and low-salt conditions (PDB entries 1ypv, 4h1i, 4gyh, 3egy and 3ehi). Here, the structure of apo hTS crystallized in the active form with sulfate ions coordinated by the arginine residue that binds dUMP is reported.
- Laboratoire de Cristallographie et RMN Biologiques, UMR CNRS 8015, Université Paris Descartes, Sorbonne Paris Cité, Faculté de Pharmacie de Paris, Paris, France.
Organizational Affiliation: