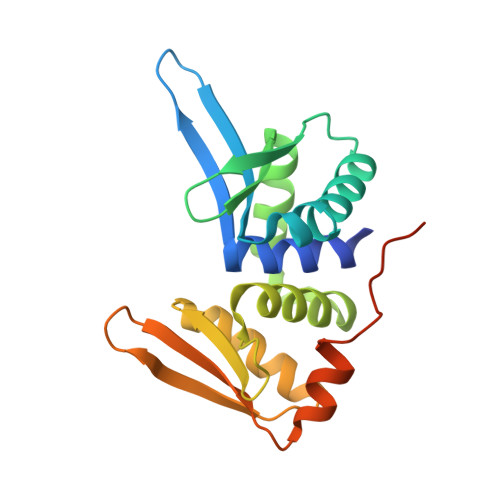
Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Molina, R., Stella, S., Redondo, P., Gomez, H., Marcaida, M.J., Orozco, M., Prieto, J., Montoya, G.(2015) Nat Struct Mol Biol 22: 65
- PubMed: 25486305
- DOI: https://doi.org/10.1038/nsmb.2932
- Primary Citation of Related Structures:
4D6N, 4D6O, 4UN7, 4UN8, 4UN9, 4UNA, 4UNB, 4UNC, 4UT0 - PubMed Abstract:
The enzymatic hydrolysis of DNA phosphodiester bonds has been widely studied, but the chemical reaction has not yet been observed. Here we follow the generation of a DNA double-strand break (DSB) by the Desulfurococcus mobilis homing endonuclease I-DmoI, trapping sequential stages of a two-metal-ion cleavage mechanism. We captured intermediates of the different catalytic steps, and this allowed us to watch the reaction by 'freezing' multiple states. We observed the successive entry of two metals involved in the reaction and the arrival of a third cation in a central position of the active site. This third metal ion has a crucial role, triggering the consecutive hydrolysis of the targeted phosphodiester bonds in the DNA strands and leaving its position once the DSB is generated. The multiple structures show the orchestrated conformational changes in the protein residues, nucleotides and metals during catalysis.
- Macromolecular Crystallography Group, Structural Biology and Biocomputing Programme, Spanish National Cancer Research Centre (CNIO), Madrid, Spain.
Organizational Affiliation: