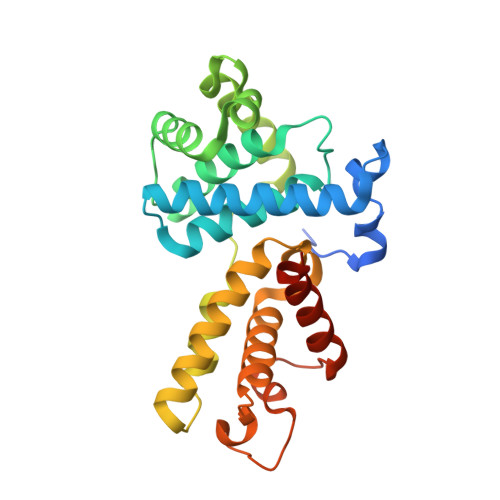
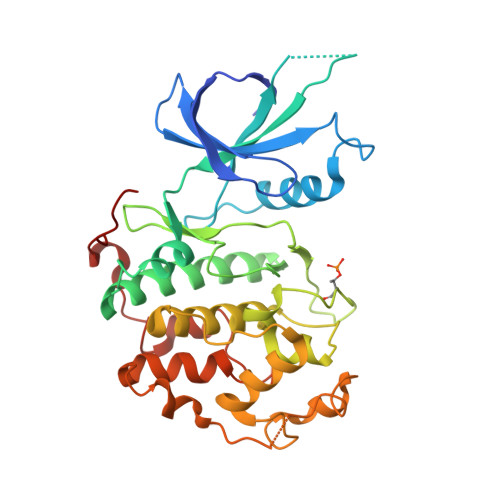
Structures of the Cdk12/Cyck Complex with AMP-Pnp Reveal a Flexible C-Terminal Kinase Extension Important for ATP Binding.
Dixon-Clarke, S.E., Elkins, J.M., Cheng, S.G., Morin, G.B., Bullock, A.N.(2015) Sci Rep 5: 17122
- PubMed: 26597175
- DOI: https://doi.org/10.1038/srep17122
- Primary Citation of Related Structures:
4CXA, 4UN0 - PubMed Abstract:
Cyclin-dependent kinase 12 (CDK12) promotes transcriptional elongation by phosphorylation of the RNA polymerase II C-terminal domain (CTD). Structure-function studies show that this activity is dependent on a C-terminal kinase extension, as well as the binding of cyclin K (CycK). To better define these interactions we determined the crystal structure of the human CDK12/CycK complex with and without the kinase extension in the presence of AMP-PNP. The structures revealed novel features for a CDK, including a large β4-β5 loop insertion that contributes to the N-lobe interaction with the cyclin. We also observed two different conformations of the C-terminal kinase extension that effectively open and close the ATP pocket. Most notably, bound AMP-PNP was only observed when trapped in the closed state. Truncation of this C-terminal structure also diminished AMP-PNP binding, as well as the catalytic activity of the CDK12/CycK complex. Further kinetic measurements showed that the full length CDK12/CycK complex was significantly more active than the two crystallised constructs suggesting a critical role for additional domains. Overall, these results demonstrate the intrinsic flexibility of the C-terminal extension in CDK12 and highlight its importance for both ATP binding and kinase activity.
- Structural Genomics Consortium, University of Oxford, Old Road Campus, Roosevelt Drive, Oxford OX3 7DQ, UK.
Organizational Affiliation: