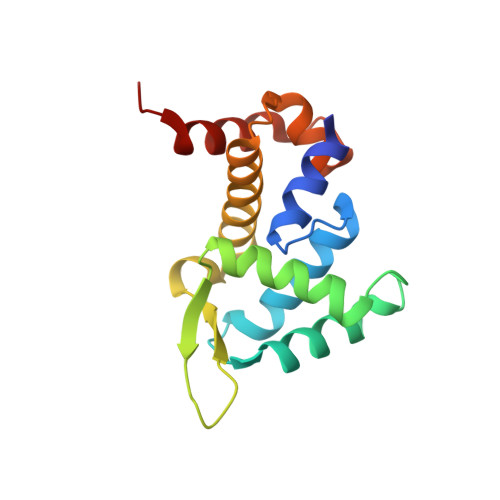
Roquin Binding to Target Mrnas Involves a Winged Helix-Turn- Helix Motif.
Schuetz, A., Murakawa, Y., Rosenbaum, E., Landthaler, M., Heinemann, U.(2014) Nat Commun 5: 5701
- PubMed: 25504471
- DOI: https://doi.org/10.1038/ncomms6701
- Primary Citation of Related Structures:
4ULW - PubMed Abstract:
Roquin proteins mediate mRNA deadenylation by recognizing a conserved class of stem-loop RNA degradation motifs via their Roquin domain. Here we present the crystal structure of a Roquin domain, revealing a mostly helical protein fold bearing a winged helix-turn-helix motif. By combining structural, biochemical and mutation analyses, we gain insight into the mode of RNA binding. We show that the winged helix-turn-helix motif is involved in the binding of constitutive decay elements-containing stem-loop mRNAs. Moreover, we provide biochemical evidence that Roquin proteins are additionally able to bind to duplex RNA and have the potential to be functional in different oligomeric states.
- Helmholtz Protein Sample Production Facility, Max Delbrück Center for Molecular Medicine, 13092 Berlin, Germany.
Organizational Affiliation: