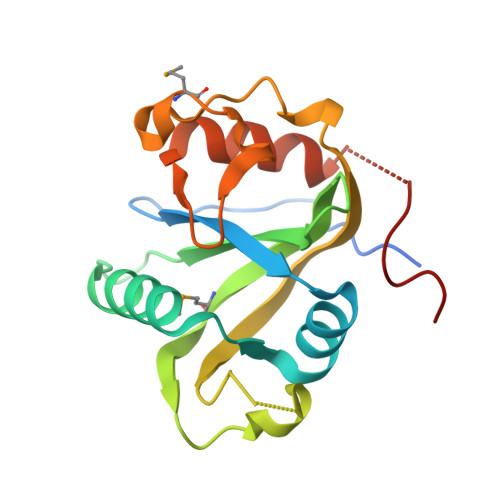
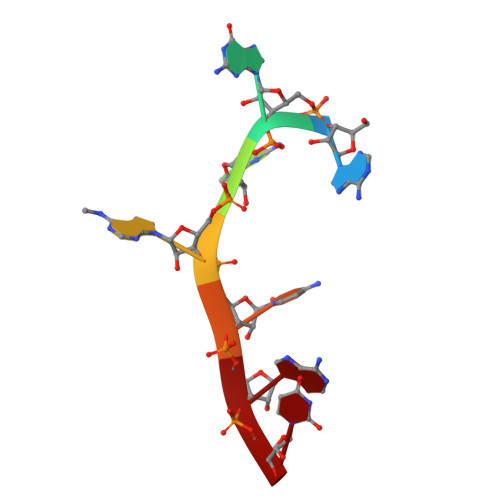
Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain.
Luo, S., Tong, L.(2014) Proc Natl Acad Sci U S A 111: 13834-13839
- PubMed: 25201973
- DOI: https://doi.org/10.1073/pnas.1412742111
- Primary Citation of Related Structures:
4U8T - PubMed Abstract:
Methylation of the N6 position of selected internal adenines (m(6)A) in mRNAs and noncoding RNAs is widespread in eukaryotes, and the YTH domain in a collection of proteins recognizes this modification. We report the crystal structure of the splicing factor YT521-B homology (YTH) domain of Zygosaccharomyces rouxii MRB1 in complex with a heptaribonucleotide with an m(6)A residue in the center. The m(6)A modification is recognized by an aromatic cage, being sandwiched between a Trp and Tyr residue and with the methyl group pointed toward another Trp residue. Mutations of YTH domain residues in the RNA binding site can abolish the formation of the complex, confirming the structural observations. These residues are conserved in the human YTH proteins that also bind m(6)A RNA, suggesting a conserved mode of recognition. Overall, our structural and biochemical studies have defined the molecular basis for how the YTH domain functions as a reader of methylated adenines.
- Department of Biological Sciences, Columbia University, New York, NY 10027.
Organizational Affiliation: