Naturally Processed Non-canonical HLA-A*02:01 Presented Peptides.
Hassan, C., Chabrol, E., Jahn, L., Kester, M.G., de Ru, A.H., Drijfhout, J.W., Rossjohn, J., Falkenburg, J.H., Heemskerk, M.H., Gras, S., van Veelen, P.A.(2015) J Biological Chem 290: 2593-2603
- PubMed: 25505266
- DOI: https://doi.org/10.1074/jbc.M114.607028
- Primary Citation of Related Structures:
4U6X, 4U6Y - PubMed Abstract:
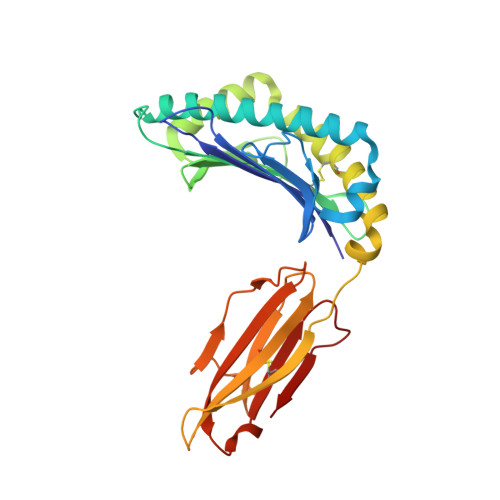
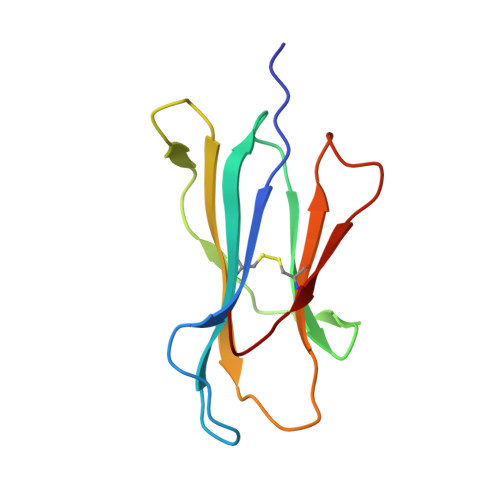
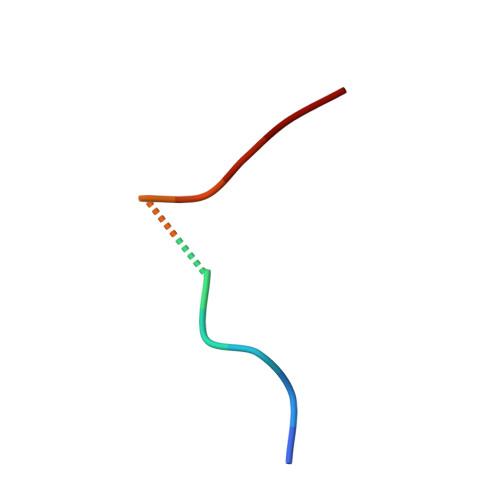
Human leukocyte antigen (HLA) class I molecules generally present peptides (p) of 8 to 11 amino acids (aa) in length. Although an increasing number of examples with lengthy (>11 aa) peptides, presented mostly by HLA-B alleles, have been reported. Here we characterize HLA-A*02:01 restricted, in addition to the HLA-B*0702 and HLA-B*4402 restricted, lengthy peptides (>11 aa) arising from the B-cell ligandome. We analyzed a number of 15-mer peptides presented by HLA-A*02:01, and confirmed pHLA-I formation by HLA folding and thermal stability assays. Surprisingly the binding affinity and stability of the 15-mer epitopes in complex with HLA-A*02:01 were comparable with the values observed for canonical length (8 to 11 aa) HLA-A*02:01-restricted peptides. We solved the structures of two 15-mer epitopes in complex with HLA-A*02:01, within which the peptides adopted distinct super-bulged conformations. Moreover, we demonstrate that T-cells can recognize the 15-mer peptides in the context of HLA-A*02:01, indicating that these 15-mer peptides represent immunogenic ligands. Collectively, our data expand our understanding of longer epitopes in the context of HLA-I, highlighting that they are not limited to the HLA-B family, but can bind the ubiquitous HLA-A*02:01 molecule, and play an important role in T-cell immunity.
- From the Departments of Immunohematology and Blood Transfusion and.
Organizational Affiliation: