Structural Fidelity and Neutralizing Antibody Binding Capacity of Hydrocarbon-Stapled HIV-1 Antigens
Bird, G.H., Irimia, A., Ofek, G., Kwong, P.D., Wilson, I.A., Walensky, L.D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 10E8 Fab Heavy Chain | A [auth H], C [auth A] | 236 | Homo sapiens | Mutation(s): 0 | 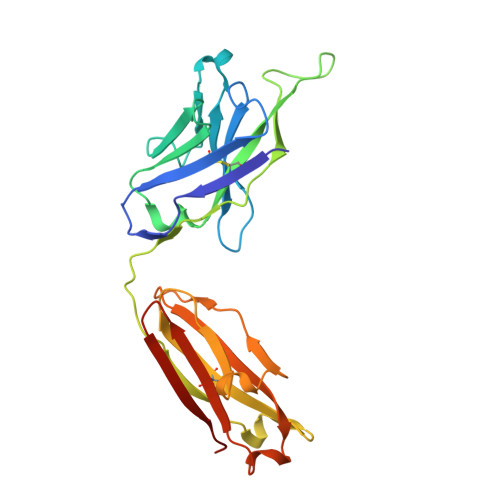 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 10E8 Fab Light Chain | B [auth L], D [auth B] | 215 | Homo sapiens | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SAH-MPER(662-683KKK)(B,q) | E [auth P], F [auth C] | 17 | Human immunodeficiency virus 1 | Mutation(s): 0 | 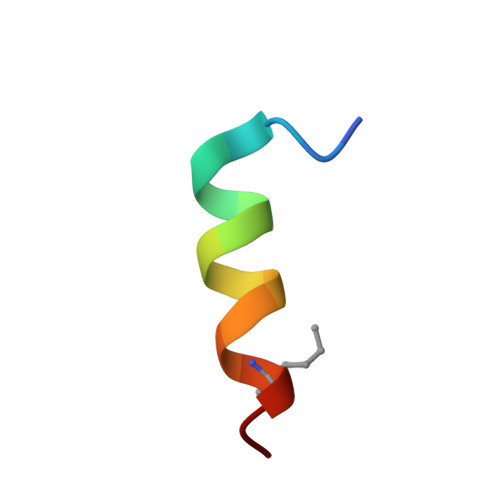 |
UniProt | |||||
Find proteins for I3VHW6 (Human immunodeficiency virus type 1) Explore I3VHW6 Go to UniProtKB: I3VHW6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | I3VHW6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MK8 Query on MK8 | E [auth P], F [auth C] | L-PEPTIDE LINKING | C7 H15 N O2 |  | LEU |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 68.562 | α = 90 |
| b = 126.317 | β = 90 |
| c = 129.875 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| PDB_EXTRACT | data extraction |
| DENZO | data reduction |
| SCALEPACK | data scaling |