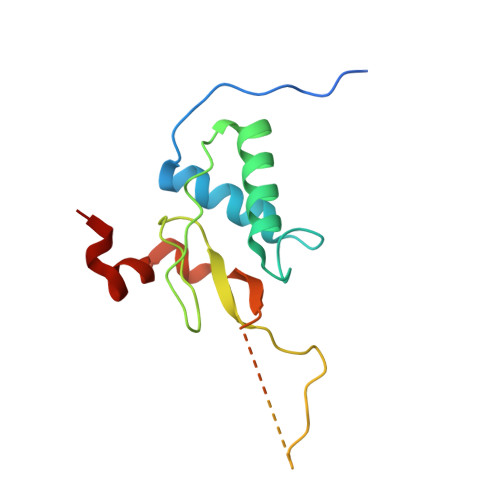
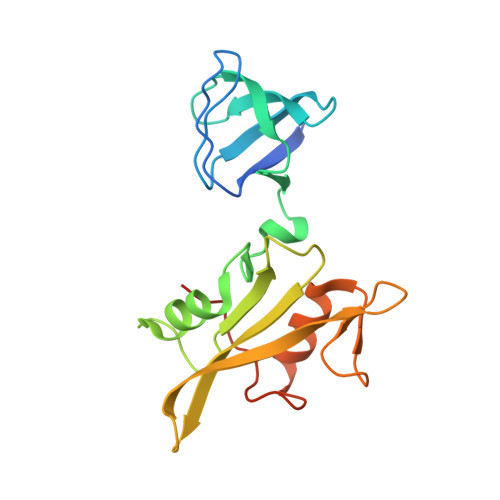
Interaction with the Src Homology (SH3-SH2) Region of the Src-family Kinase Hck Structures the HIV-1 Nef Dimer for Kinase Activation and Effector Recruitment.
Alvarado, J.J., Tarafdar, S., Yeh, J.I., Smithgall, T.E.(2014) J Biological Chem 289: 28539-28553
- PubMed: 25122770
- DOI: https://doi.org/10.1074/jbc.M114.600031
- Primary Citation of Related Structures:
4U5W - PubMed Abstract:
HIV-1 Nef supports high titer viral replication in vivo and is essential for AIDS progression. Nef function depends on interactions with multiple host cell effectors, including Hck and other Src-family kinases. Here we describe the x-ray crystal structure of Nef in complex with the Hck SH3-SH2 regulatory region to a resolution of 1.86 Å. The complex crystallized as a dimer of complexes, with the conserved Nef PXXPXR motif engaging the Hck SH3 domain. A new intercomplex contact was found between SH3 Glu-93, and Nef Arg-105. Mutagenesis of Hck SH3 Glu-93 interfered with Nef·Hck complex formation and kinase activation in cells. The Hck SH2 domains impinge on the N-terminal region of Nef to stabilize a dimer conformation that exposes Asp-123, a residue critical for Nef function. Our results suggest that in addition to serving as a kinase effector for Nef, Hck binding may reorganize the Nef dimer for functional interaction with other signaling partners.
- From the Departments of Microbiology and Molecular Genetics and Structural Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 15219 and.
Organizational Affiliation: