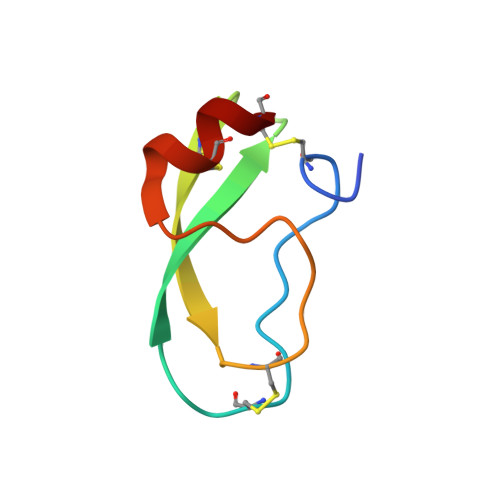
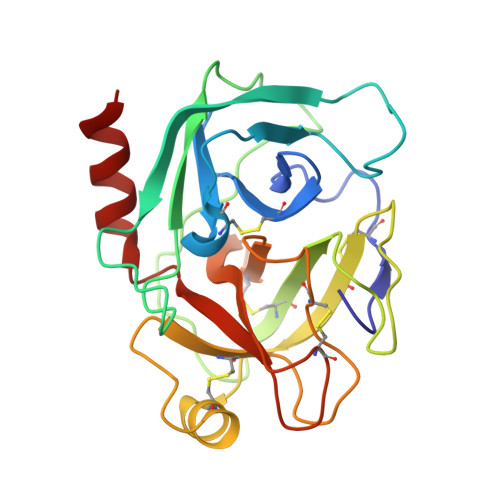
Sequence and Conformational Specificity in Substrate Recognition: SEVERAL HUMAN KUNITZ PROTEASE INHIBITOR DOMAINS ARE SPECIFIC SUBSTRATES OF MESOTRYPSIN.
Pendlebury, D., Wang, R., Henin, R.D., Hockla, A., Soares, A.S., Madden, B.J., Kazanov, M.D., Radisky, E.S.(2014) J Biological Chem 289: 32783-32797
- PubMed: 25301953
- DOI: https://doi.org/10.1074/jbc.M114.609560
- Primary Citation of Related Structures:
4U30, 4U32 - PubMed Abstract:
Mesotrypsin is an isoform of trypsin that is uniquely resistant to polypeptide trypsin inhibitors and can cleave some inhibitors rapidly. Previous studies have shown that the amyloid precursor protein Kunitz protease inhibitor domain (APPI) is a specific substrate of mesotrypsin and that stabilization of the APPI cleavage site in a canonical conformation contributes to recognition by mesotrypsin. We hypothesized that other proteins possessing potential cleavage sites stabilized in a similar conformation might also be mesotrypsin substrates. Here we evaluated a series of candidate substrates, including human Kunitz protease inhibitor domains from amyloid precursor-like protein 2 (APLP2), bikunin, hepatocyte growth factor activator inhibitor type 2 (HAI2), tissue factor pathway inhibitor-1 (TFPI1), and tissue factor pathway inhibitor-2 (TFPI2), as well as E-selectin, an unrelated protein possessing a potential cleavage site displaying canonical conformation. We find that Kunitz domains within APLP2, bikunin, and HAI2 are cleaved by mesotrypsin with kinetic profiles of specific substrates. TFPI1 and TFPI2 Kunitz domains are cleaved less efficiently by mesotrypsin, and E-selectin is not cleaved at the anticipated site. Cocrystal structures of mesotrypsin with HAI2 and bikunin Kunitz domains reveal the mode of mesotrypsin interaction with its canonical substrates. Our data suggest that major determinants of mesotrypsin substrate specificity include sequence preferences at the P1 and P'2 positions along with conformational stabilization of the cleavage site in the canonical conformation. Mesotrypsin up-regulation has been implicated previously in cancer progression, and proteolytic clearance of Kunitz protease inhibitors offers potential mechanisms by which mesotrypsin may mediate pathological effects in cancer.
- From the Department of Cancer Biology, Mayo Clinic Comprehensive Cancer Center, Jacksonville, Florida 32224.
Organizational Affiliation: