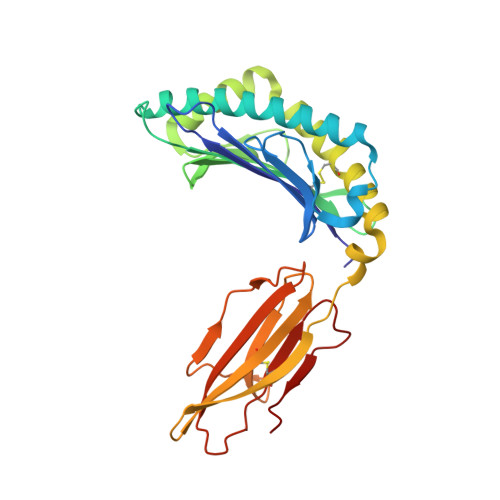
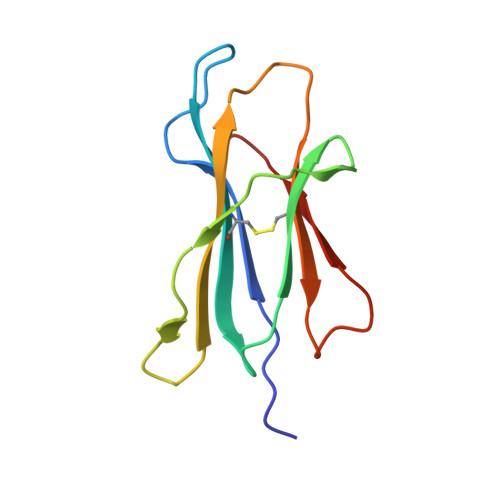
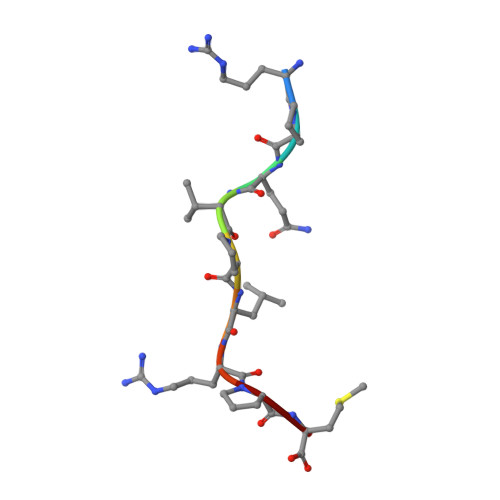
A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Klverpris, H.N., Cole, D.K., Fuller, A., Carlson, J., Beck, K., Schauenburg, A.J., Rizkallah, P.J., Buus, S., Sewell, A.K., Goulder, P.(2015) Retrovirology 12: 20-20
- PubMed: 25808313
- DOI: https://doi.org/10.1186/s12977-015-0149-5
- Primary Citation of Related Structures:
4U1H, 4U1I, 4U1J, 4U1K, 4U1L, 4U1M, 4U1N, 4U1S - PubMed Abstract:
Presentation of identical HIV-1 peptides by closely related Human Leukocyte Antigen class I (HLAI) molecules can select distinct patterns of escape mutation that have a significant impact on viral fitness and disease progression. The molecular mechanisms by which HLAI micropolymorphisms can induce differential HIV-1 escape patterns within identical peptide epitopes remain unknown. Here, we undertook genetic and structural analyses of two immunodominant HIV-1 peptides, Gag180-188 (TPQDLNTML, TL9-p24) and Nef71-79 (RPQVPLRPM, RM9-Nef) that are among the most highly targeted epitopes in the global HIV-1 epidemic. We show that single polymorphisms between different alleles of the HLA-B7 superfamily can induce a conformational switch in peptide conformation that is associated with differential HLAI-specific escape mutation and immune control. A dominant R71K mutation in the Nef71-79 occurred in those with HLA-B*07:02 but not B*42:01/02 or B*81:01. No structural difference in the HLA-epitope complexes was detected to explain this observation. These data suggest that identical peptides presented through very similar HLAI landscapes are recognized as distinct epitopes and provide a novel structural mechanism for previously observed differential HIV-1 escape and disease progression.