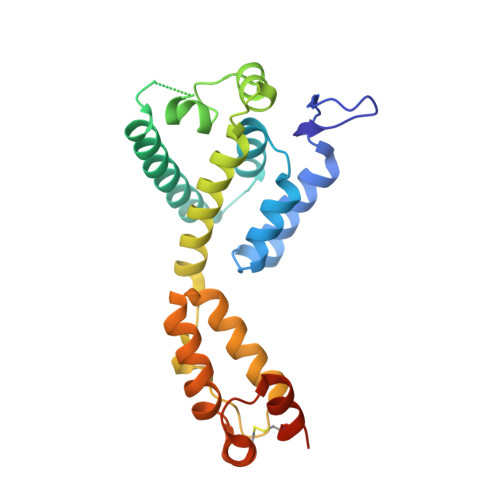
Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Price, A.J., Jacques, D.A., McEwan, W.A., Fletcher, A.J., Essig, S., Chin, J.W., Halambage, U.D., Aiken, C., James, L.C.(2014) PLoS Pathog 10: e1004459-e1004459
- PubMed: 25356722
- DOI: https://doi.org/10.1371/journal.ppat.1004459
- Primary Citation of Related Structures:
4U0A, 4U0B, 4U0C, 4U0D, 4U0E, 4U0F - PubMed Abstract:
The HIV-1 capsid is involved in all infectious steps from reverse transcription to integration site selection, and is the target of multiple host cell and pharmacologic ligands. However, structural studies have been limited to capsid monomers (CA), and the mechanistic basis for how these ligands influence infection is not well understood. Here we show that a multi-subunit interface formed exclusively within CA hexamers mediates binding to linear epitopes within cellular cofactors NUP153 and CPSF6, and is competed for by the antiretroviral compounds PF74 and BI-2. Each ligand is anchored via a shared phenylalanine-glycine (FG) motif to a pocket within the N-terminal domain of one monomer, and all but BI-2 also make essential interactions across the N-terminal domain: C-terminal domain (NTD:CTD) interface to a second monomer. Dissociation of hexamer into CA monomers prevents high affinity interaction with CPSF6 and PF74, and abolishes binding to NUP153. The second interface is conformationally dynamic, but binding of NUP153 or CPSF6 peptides is accommodated by only one conformation. NUP153 and CPSF6 have overlapping binding sites, but each makes unique CA interactions that, when mutated selectively, perturb cofactor dependency. These results reveal that multiple ligands share an overlapping interface in HIV-1 capsid that is lost upon viral disassembly.
- Medical Research Council Laboratory of Molecular Biology, Division of Protein and Nucleic Acid Chemistry, Cambridge, United Kingdom.
Organizational Affiliation: