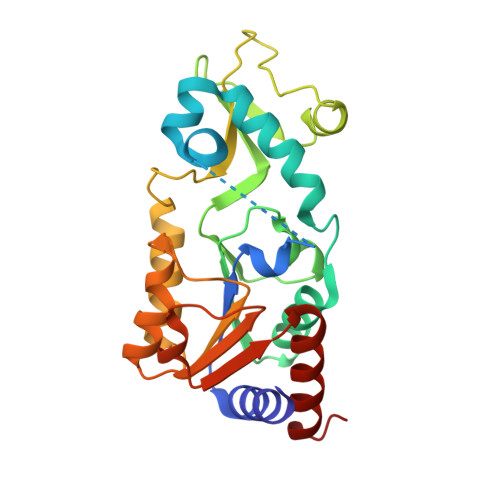
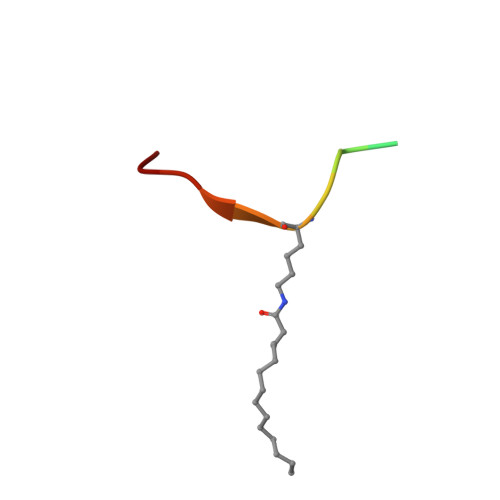
Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Ringel, A.E., Roman, C., Wolberger, C.(2014) Protein Sci 23: 1686-1697
- PubMed: 25200501
- DOI: https://doi.org/10.1002/pro.2546
- Primary Citation of Related Structures:
4TWI, 4TWJ - PubMed Abstract:
Sirtuins were originally shown to regulate a wide array of biological processes such as transcription, genomic stability, and metabolism by catalyzing the NAD(+) -dependent deacetylation of lysine residues. Recent proteomic studies have revealed a much wider array of lysine acyl modifications in vivo than was previously known, which has prompted a reevaluation of sirtuin substrate specificity. Several sirtuins have now been shown to preferentially remove propionyl, succinyl, and long-chain fatty acyl groups from lysines, which has changed our understanding of sirtuin biology. In light of these developments, we revisited the acyl specificity of several well-studied archaeal and bacterial sirtuins. We find that the Archaeoglobus fulgidus sirtuins, Sir2Af1 and Sir2Af2, preferentially remove succinyl and myristoyl groups, respectively. Crystal structures of Sir2Af1 bound to a succinylated peptide and Sir2Af2 bound to a myristoylated peptide show how the active site of each enzyme accommodates a noncanonical acyl chain. As compared to its structure in complex with an acetylated peptide, Sir2Af2 undergoes a conformational change that expands the active site to accommodate the myristoyl group. These findings point to both structural and biochemical plasticity in sirtuin active sites and provide further evidence that sirtuins from all three domains of life catalyze noncanonical deacylation.
- Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, Maryland, 21205-2185.
Organizational Affiliation: