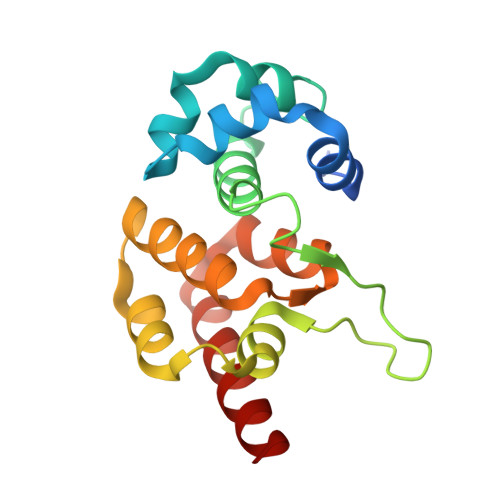
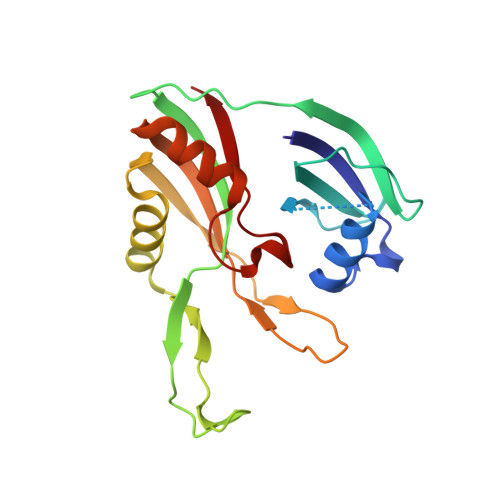
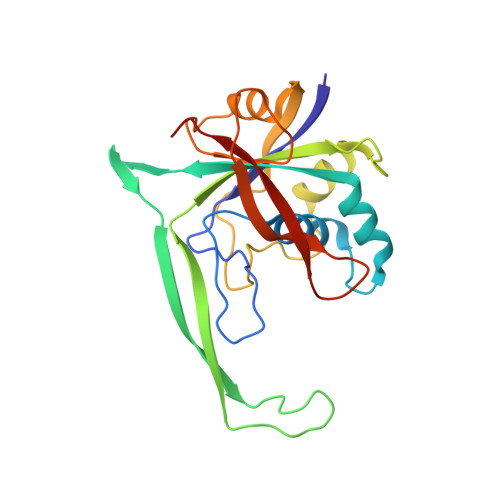
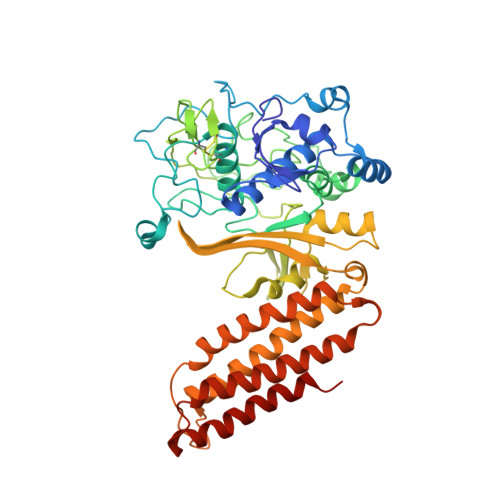
Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli.
Jackson, R.N., Golden, S.M., van Erp, P.B., Carter, J., Westra, E.R., Brouns, S.J., van der Oost, J., Terwilliger, T.C., Read, R.J., Wiedenheft, B.(2014) Science 345: 1473-1479
- PubMed: 25103409
- DOI: https://doi.org/10.1126/science.1256328
- Primary Citation of Related Structures:
4TVX - PubMed Abstract:
Clustered regularly interspaced short palindromic repeats (CRISPRs) are essential components of RNA-guided adaptive immune systems that protect bacteria and archaea from viruses and plasmids. In Escherichia coli, short CRISPR-derived RNAs (crRNAs) assemble into a 405-kilodalton multisubunit surveillance complex called Cascade (CRISPR-associated complex for antiviral defense). Here we present the 3.24 angstrom resolution x-ray crystal structure of Cascade. Eleven proteins and a 61-nucleotide crRNA assemble into a seahorse-shaped architecture that binds double-stranded DNA targets complementary to the crRNA-guide sequence. Conserved sequences on the 3' and 5' ends of the crRNA are anchored by proteins at opposite ends of the complex, whereas the guide sequence is displayed along a helical assembly of six interwoven subunits that present five-nucleotide segments of the crRNA in pseudo-A-form configuration. The structure of Cascade suggests a mechanism for assembly and provides insights into the mechanisms of target recognition.
- Department of Microbiology and Immunology, Montana State University, Bozeman, MT 59717, USA.
Organizational Affiliation: