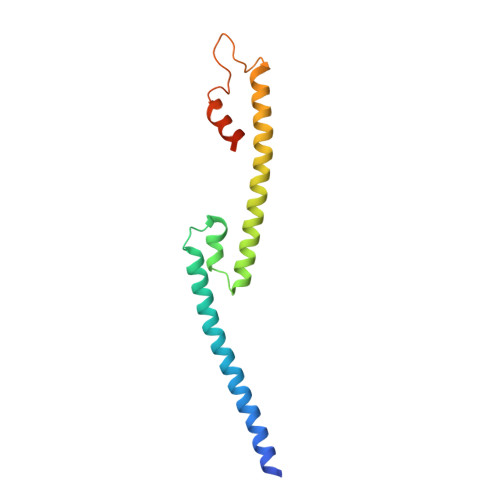
Insights into Herpesvirus Tegument Organization from Structural Analyses of the 970 Central Residues of HSV-1 UL36 Protein.
Scrima, N., Lepault, J., Boulard, Y., Pasdeloup, D., Bressanelli, S., Roche, S.(2015) J Biological Chem 290: 8820-8833
- PubMed: 25678705
- DOI: https://doi.org/10.1074/jbc.M114.612838
- Primary Citation of Related Structures:
4TT0, 4TT1 - PubMed Abstract:
The tegument of all herpesviruses contains a capsid-bound large protein that is essential for multiple viral processes, including capsid transport, decapsidation at the nuclear pore complex, particle assembly, and secondary envelopment, through mechanisms that are still incompletely understood. We report here a structural characterization of the central 970 residues of this protein for herpes simplex virus type 1 (HSV-1 UL36, 3164 residues). This large fragment is essentially a 34-nm-long monomeric fiber. The crystal structure of its C terminus shows an elongated domain-swapped dimer. Modeling and molecular dynamics simulations give a likely molecular organization for the monomeric form and extend our findings to alphaherpesvirinae. Hence, we propose that an essential feature of UL36 is the existence in its central region of a stalk capable of connecting capsid and membrane across the tegument and that the ability to switch between monomeric and dimeric forms may help UL36 fulfill its multiple functions.
- From the Institute for Integrative Biology of the Cell (I2BC), 1 Avenue de la Terrasse, 91198 Gif-sur-Yvette.
Organizational Affiliation: