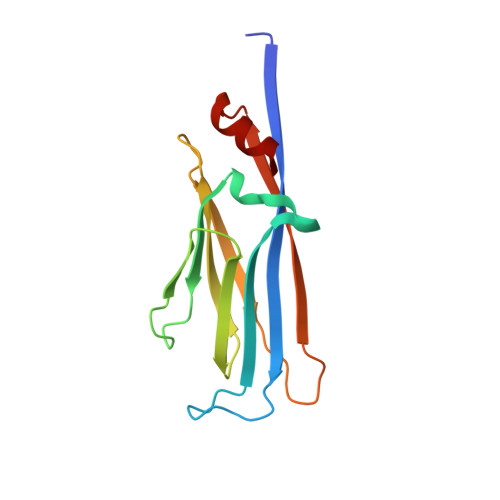AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation.
Li, Y., Wen, H., Xi, Y., Tanaka, K., Wang, H., Peng, D., Ren, Y., Jin, Q., Dent, S.Y., Li, W., Li, H., Shi, X.(2014) Cell 159: 558-571
- PubMed: 25417107
- DOI: https://doi.org/10.1016/j.cell.2014.09.049
- Primary Citation of Related Structures:
4TMP - PubMed Abstract:
The recognition of modified histones by "reader" proteins constitutes a key mechanism regulating gene expression in the chromatin context. Compared with the great variety of readers for histone methylation, few protein modules that recognize histone acetylation are known. Here, we show that the AF9 YEATS domain binds strongly to histone H3K9 acetylation and, to a lesser extent, H3K27 and H3K18 acetylation. Crystal structural studies revealed that AF9 YEATS adopts an eight-stranded immunoglobin fold and utilizes a serine-lined aromatic "sandwiching" cage for acetyllysine readout, representing a novel recognition mechanism that is distinct from that of known acetyllysine readers. ChIP-seq experiments revealed a strong colocalization of AF9 and H3K9 acetylation genome-wide, which is important for the chromatin recruitment of the H3K79 methyltransferase DOT1L. Together, our studies identified the evolutionarily conserved YEATS domain as a novel acetyllysine-binding module and established a direct link between histone acetylation and DOT1L-mediated H3K79 methylation in transcription control.
- Collaborative Innovation Center for Biotherapy, MOE Key Laboratory of Protein Sciences, Center for Structural Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China; Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China; Tsinghua-Peking Center for Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: