Peptide Inhibitors of the Escherichia coli DsbA Oxidative Machinery Essential for Bacterial Virulence.
Duprez, W., Premkumar, L., Halili, M.A., Lindahl, F., Reid, R.C., Fairlie, D.P., Martin, J.L.(2015) J Med Chem 58: 577-587
- PubMed: 25470204
- DOI: https://doi.org/10.1021/jm500955s
- Primary Citation of Related Structures:
4TKY - PubMed Abstract:
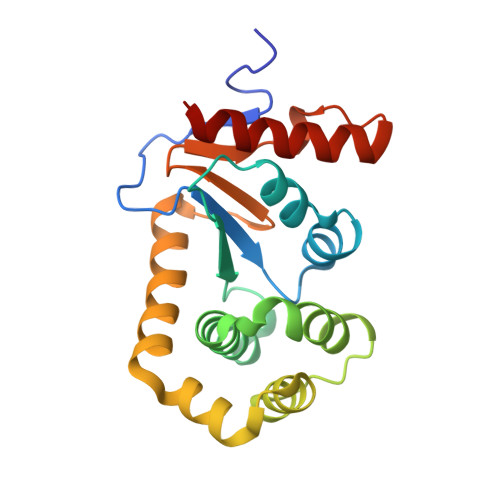
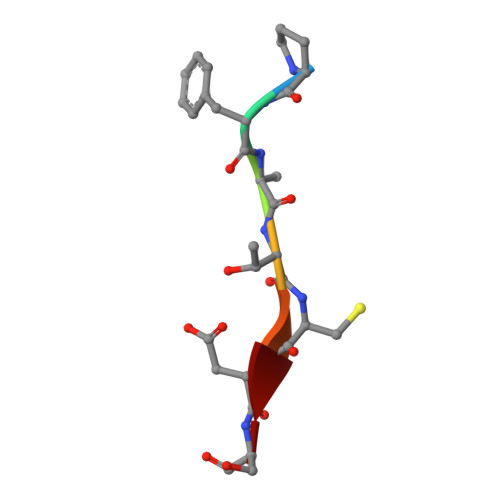
One approach to address antibiotic resistance is to develop drugs that interfere with bacterial virulence. A master regulator of virulence in Gram-negative bacteria is the oxidative folding machinery comprising DsbA and DsbB. A crystal structure at 2.5 Å resolution is reported here for Escherichia coli DsbA complexed with PFATCDS, a heptapeptide derived from the partner protein Escherichia coli DsbB. Details of the peptide binding mode and binding site provide valuable clues for inhibitor design. Structure-activity relationships for 30 analogues were used to produce short peptides with a cysteine that bind tightly to EcDsbA (Kd = 2.0 ± 0.3 μM) and inhibit its activity (IC50 = 5.1 ± 1.1 μM). The most potent inhibitor does not bind to or inhibit human thioredoxin that shares a similar active site. This finding suggests that small molecule inhibitors can be designed to exploit a key interaction of EcDsbA, as the basis for antivirulence agents with a novel mechanism of action.
- Institute for Molecular Bioscience, Division of Chemistry and Structural Biology, University of Queensland , Brisbane, Queensland 4072, Australia.
Organizational Affiliation: