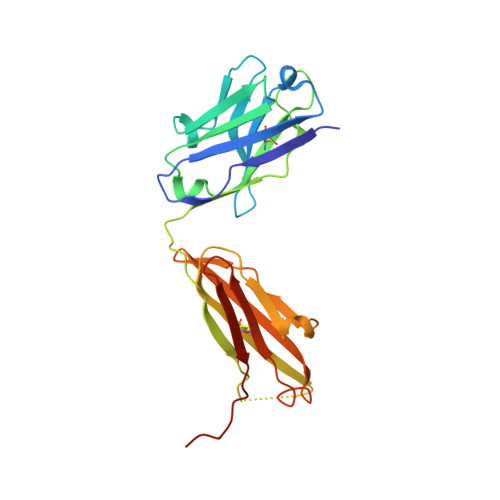
A Linear Epitope in the N-Terminal Domain of CCR5 and Its Interaction with Antibody.
Chain, B., Arnold, J., Akthar, S., Brandt, M., Davis, D., Noursadeghi, M., Lapp, T., Ji, C., Sankuratri, S., Zhang, Y., Govada, L., Saridakis, E., Chayen, N.(2015) PLoS One 10: e0128381-e0128381
- PubMed: 26030924
- DOI: https://doi.org/10.1371/journal.pone.0128381
- Primary Citation of Related Structures:
4S2S - PubMed Abstract:
The CCR5 receptor plays a role in several key physiological and pathological processes and is an important therapeutic target. Inhibition of the CCR5 axis by passive or active immunisation offers one very selective strategy for intervention. In this study we define a new linear epitope within the extracellular domain of CCR5 recognised by two independently produced monoclonal antibodies. A short peptide encoding the linear epitope can induce antibodies which recognise the intact receptor when administered colinear with a tetanus toxoid helper T cell epitope. The monoclonal antibody RoAb 13 is shown to bind to both cells and peptide with moderate to high affinity (6x10^8 and 1.2x107 M-1 respectively), and binding to the peptide is enhanced by sulfation of tyrosines at positions 10 and 14. RoAb13, which has previously been shown to block HIV infection, also blocks migration of monocytes in response to CCR5 binding chemokines and to inflammatory macrophage conditioned medium. A Fab fragment of RoAb13 has been crystallised and a structure of the antibody is reported to 2.1 angstrom resolution.
- Division of Infection and Immunity, UCL, Gower St., London, United Kingdom.
Organizational Affiliation: