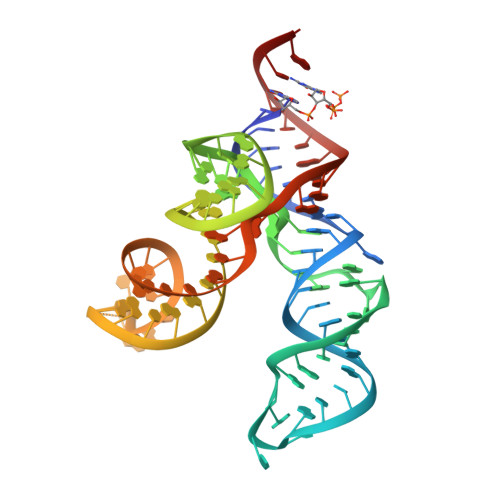
Bacterial riboswitches cooperatively bind Ni(2+) or Co(2+) ions and control expression of heavy metal transporters.
Furukawa, K., Ramesh, A., Zhou, Z., Weinberg, Z., Vallery, T., Winkler, W.C., Breaker, R.R.(2015) Mol Cell 57: 1088-1098
- PubMed: 25794617
- DOI: https://doi.org/10.1016/j.molcel.2015.02.009
- Primary Citation of Related Structures:
4RUM - PubMed Abstract:
Bacteria regularly encounter widely varying metal concentrations in their surrounding environment. As metals become depleted or, conversely, accrue to toxicity, microbes will activate cellular responses that act to maintain metal homeostasis. A suite of metal-sensing regulatory ("metalloregulatory") proteins orchestrate these responses by allosterically coupling the selective binding of target metals to the activity of DNA-binding domains. However, we report here the discovery, validation, and structural details of a widespread class of riboswitch RNAs, whose members selectively and tightly bind the low-abundance transition metals, Ni(2+) and Co(2+). These riboswitches bind metal cooperatively, and with affinities in the low micromolar range. The structure of a Co(2+)-bound RNA reveals a network of molecular contacts that explains how it achieves cooperative binding between adjacent sites. These findings reveal that bacteria have evolved to utilize highly selective metalloregulatory riboswitches, in addition to metalloregulatory proteins, for detecting and responding to toxic levels of heavy metals.
- Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06520, USA.
Organizational Affiliation: