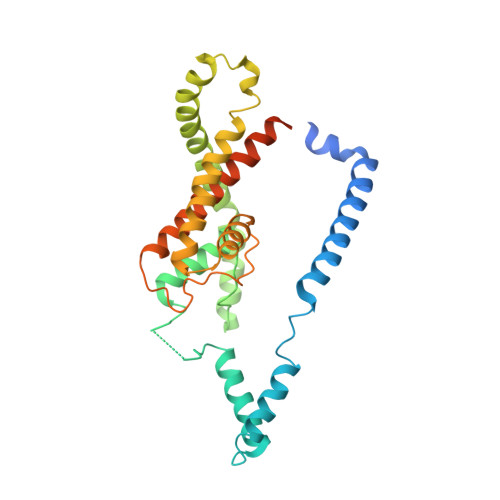
Transmembrane Helix Straightening and Buckling Underlies Activation of Mechanosensitive and Thermosensitive K2P Channels.
Lolicato, M., Riegelhaupt, P.M., Arrigoni, C., Clark, K.A., Minor, D.L.(2014) Neuron 84: 1198-1212
- PubMed: 25500157
- DOI: https://doi.org/10.1016/j.neuron.2014.11.017
- Primary Citation of Related Structures:
4RUE, 4RUF - PubMed Abstract:
Mechanical and thermal activation of ion channels is central to touch, thermosensation, and pain. The TRAAK/TREK K(2P) potassium channel subfamily produces background currents that alter neuronal excitability in response to pressure, temperature, signaling lipids, and anesthetics. How such diverse stimuli control channel function is unclear. Here we report structures of K(2P)4.1 (TRAAK) bearing C-type gate-activating mutations that reveal a tilting and straightening of the M4 inner transmembrane helix and a buckling of the M2 transmembrane helix. These conformational changes move M4 in a direction opposite to that in classical potassium channel activation mechanisms and open a passage lateral to the pore that faces the lipid bilayer inner leaflet. Together, our findings uncover a unique aspect of K(2P) modulation, indicate a means for how the K(2P) C-terminal cytoplasmic domain affects the C-type gate which lies ∼40Å away, and suggest how lipids and bilayer inner leaflet deformations may gate the channel.
- Cardiovascular Research Institute, University of California, San Francisco, San Francisco, California 93858-2330, USA.
Organizational Affiliation: