Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv
Filippova, E.V., Minasov, G., Kiryukhina, O., Endres, M., Babnigg, G., Rubin, E., Sacchettini, J., Joachimiak, A., Anderson, W.F.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chaperone protein DnaK | A [auth D] | 546 | Mycobacterium tuberculosis H37Rv | Mutation(s): 0 Gene Names: dnaK, P425_00363, RVBD_0350, WP_003401814 | 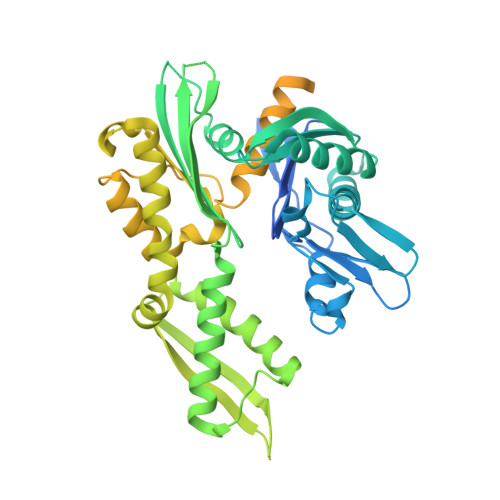 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP Query on ATP | B [auth D] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| PG4 Query on PG4 | C [auth D] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 117.866 | α = 90 |
| b = 117.866 | β = 90 |
| c = 134.943 | γ = 120 |
| Software Name | Purpose |
|---|---|
| Blu-Ice | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |