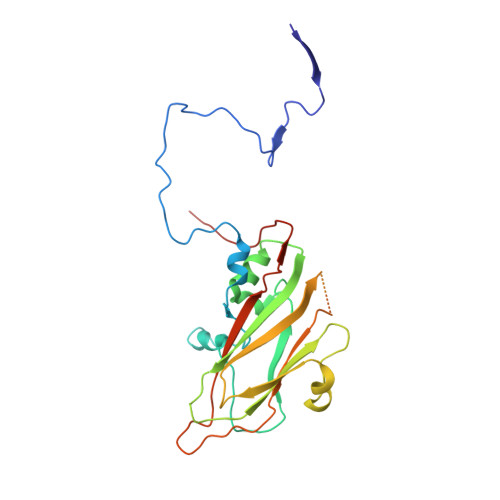
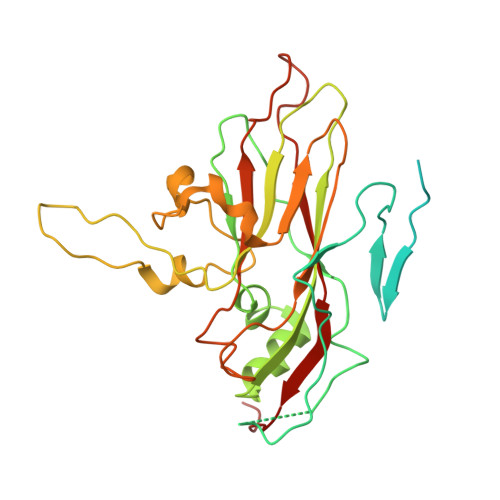
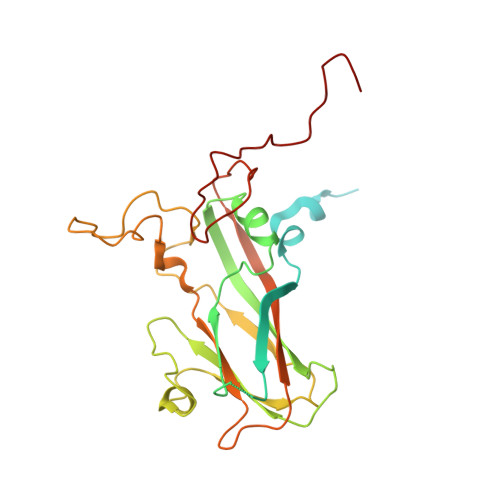
Crystal structures of enterovirus 71 (EV71) recombinant virus particles provide insights into vaccine design.
Lyu, K., Wang, G.C., He, Y.L., Han, J.F., Ye, Q., Qin, C.F., Chen, R.(2015) J Biological Chem 290: 3198-3208
- PubMed: 25492868
- DOI: https://doi.org/10.1074/jbc.M114.624536
- Primary Citation of Related Structures:
4RQP, 4RR3, 4RS5 - PubMed Abstract:
Hand-foot-and-mouth disease (HFMD) remains a major health concern in the Asia-Pacific regions, and its major causative agents include human enterovirus 71 (EV71) and coxsackievirus A16. A desirable vaccine against HFMD would be multivalent and able to elicit protective responses against multiple HFMD causative agents. Previously, we have demonstrated that a thermostable recombinant EV71 vaccine candidate can be produced by the insertion of a foreign peptide into the BC loop of VP1 without affecting viral replication. Here we present crystal structures of two different naturally occurring empty particles, one from a clinical C4 strain EV71 and the other from its recombinant virus containing an insertion in the VP1 BC loop. Crystal structure analysis demonstrated that the inserted foreign peptide is well exposed on the particle surface without significant structural changes in the capsid. Importantly, such insertions do not seem to affect the virus uncoating process as illustrated by the conformational similarity between an uncoating intermediate of another recombinant virus and that of EV71. Especially, at least 18 residues from the N terminus of VP1 are transiently externalized. Altogether, our study provides insights into vaccine development against HFMD.
- From the Key Laboratory of Molecular Virology and Immunology, Institut Pasteur of Shanghai, Chinese Academy of Sciences, Shanghai 200031, China.
Organizational Affiliation: