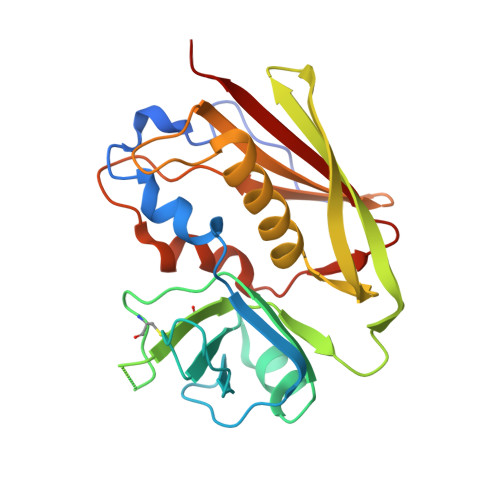
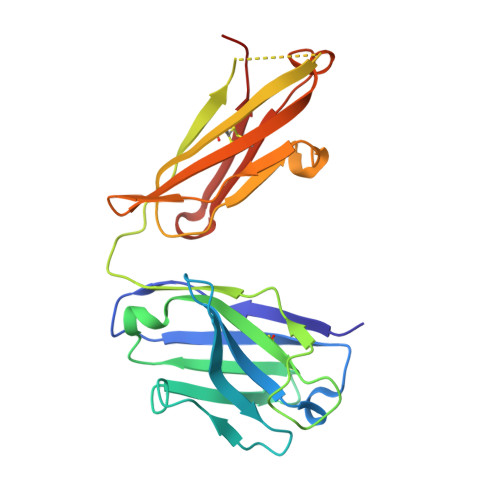
Mechanisms mediating enhanced neutralization efficacy of staphylococcal enterotoxin B by combinations of monoclonal antibodies.
Dutta, K., Varshney, A.K., Franklin, M.C., Goger, M., Wang, X., Fries, B.C.(2015) J Biological Chem 290: 6715-6730
- PubMed: 25572397
- DOI: https://doi.org/10.1074/jbc.M114.630715
- Primary Citation of Related Structures:
4RGM, 4RGN, 4RGO - PubMed Abstract:
Staphylococcal enterotoxin B (SEB) is a superantigen that cross-links the major histocompatibility complex class II and specific V-β chains of the T-cell receptor, thus forming a ternary complex. Developing neutralizing mAb to disrupt the ternary complex and abrogate the resulting toxicity is a major therapeutic challenge because SEB is effective at very low concentrations. We show that combining two SEB-specific mAbs enhances their efficacy, even though one of the two mAbs by itself has no effect on neutralization. Crystallography was employed for fine-mapping conformational epitopes in binary and ternary complexes between SEB and Fab fragments. NMR spectroscopy was used to validate and identify subtle allosteric changes induced by mAbs binding to SEB. The mapping of epitopes established that a combination of different mAbs can enhance efficacy of mAb-mediated protection from SEB induced lethal shock by two different mechanisms: one mAb mixture promoted clearance of the toxin both in vitro and in vivo by FcR-mediated cross-linking and clearance, whereas the other mAb mixture induced subtle allosteric conformational changes in SEB that perturbed formation of the SEB·T-cell receptor·major histocompatibility complex class II trimer. Finally structural information accurately predicted mAb binding to other superantigens that share conformational epitopes with SEB. Fine mapping of conformational epitopes is a powerful tool to establish the mechanism and optimize the action of synergistic mAb combinations.
- From the New York Structural Biology Center, New York, New York 10027, dutta@nysbc.org.
Organizational Affiliation: