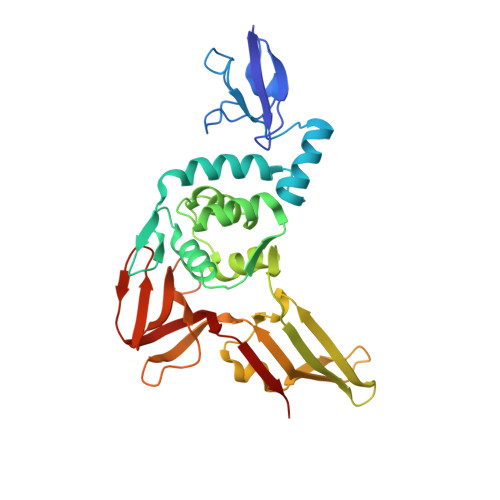
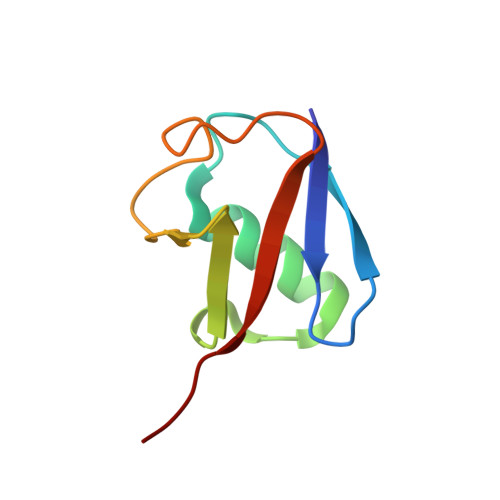
Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
Bailey-Elkin, B.A., Knaap, R.C., Johnson, G.G., Dalebout, T.J., Ninaber, D.K., van Kasteren, P.B., Bredenbeek, P.J., Snijder, E.J., Kikkert, M., Mark, B.L.(2014) J Biological Chem 289: 34667-34682
- PubMed: 25320088
- DOI: https://doi.org/10.1074/jbc.M114.609644
- Primary Citation of Related Structures:
4REZ, 4RF0, 4RF1 - PubMed Abstract:
Middle East respiratory syndrome coronavirus (MERS-CoV) is a newly emerging human pathogen that was first isolated in 2012. MERS-CoV replication depends in part on a virus-encoded papain-like protease (PL(pro)) that cleaves the viral replicase polyproteins at three sites releasing non-structural protein 1 (nsp1), nsp2, and nsp3. In addition to this replicative function, MERS-CoV PL(pro) was recently shown to be a deubiquitinating enzyme (DUB) and to possess deISGylating activity, as previously reported for other coronaviral PL(pro) domains, including that of severe acute respiratory syndrome coronavirus. These activities have been suggested to suppress host antiviral responses during infection. To understand the molecular basis for ubiquitin (Ub) recognition and deconjugation by MERS-CoV PL(pro), we determined its crystal structure in complex with Ub. Guided by this structure, mutations were introduced into PL(pro) to specifically disrupt Ub binding without affecting viral polyprotein cleavage, as determined using an in trans nsp3↓4 cleavage assay. Having developed a strategy to selectively disable PL(pro) DUB activity, we were able to specifically examine the effects of this activity on the innate immune response. Whereas the wild-type PL(pro) domain was found to suppress IFN-β promoter activation, PL(pro) variants specifically lacking DUB activity were no longer able to do so. These findings directly implicate the DUB function of PL(pro), and not its proteolytic activity per se, in the inhibition of IFN-β promoter activity. The ability to decouple the DUB activity of PL(pro) from its role in viral polyprotein processing now provides an approach to further dissect the role(s) of PL(pro) as a viral DUB during MERS-CoV infection.
- From the Department of Microbiology, University of Manitoba, Winnipeg, Manitoba R3T 2N2, Canada and.
Organizational Affiliation: