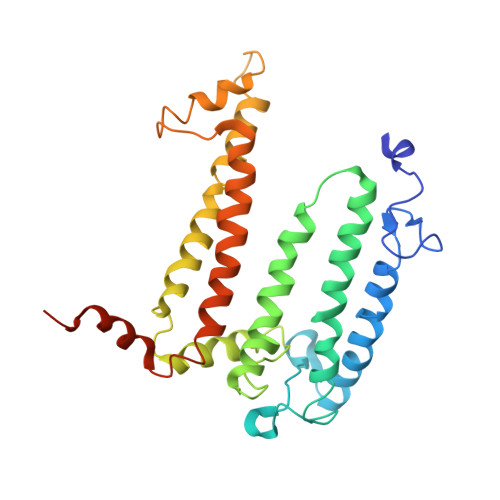
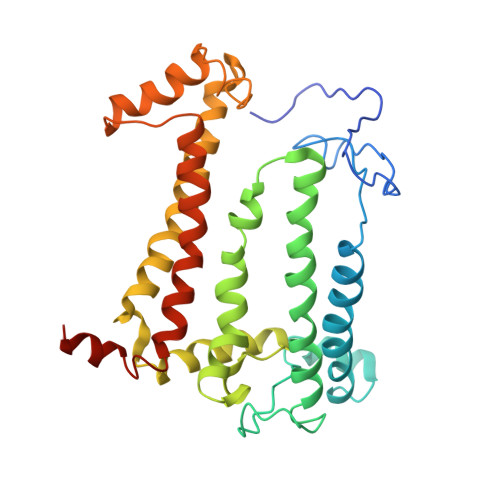
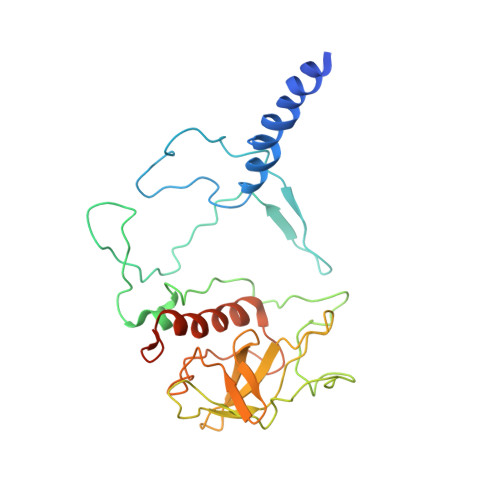
Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Yeates, T.O., Komiya, H., Chirino, A., Rees, D.C., Allen, J.P., Feher, G.(1988) Proc Natl Acad Sci U S A 85: 7993-7997
- PubMed: 3186702
- DOI: https://doi.org/10.1073/pnas.85.21.7993
- Primary Citation of Related Structures:
4RCR - PubMed Abstract:
The three-dimensional structures of the cofactors and protein subunits of the reaction center (RC) from the carotenoidless mutant strain of Rhodobacter sphaeroides R-26 and the wild-type strain 2.4.1 have been determined by x-ray diffraction to resolutions of 2.8 A and 3.0 A with R values of 24% and 26%, respectively. The bacteriochlorophyll dimer (D), bacteriochlorophyll monomers (B), and bacteriopheophytin monomers (phi) form two branches, A and B, that are approximately related by a twofold symmetry axis. The cofactors are located in hydrophobic environments formed by the L and M subunits. Differences in the cofactor-protein interactions between the A and B cofactors, as well as between the corresponding cofactors of Rb, sphaeroides and Rhodopseudomonas viridis [Michel, H., Epp, O. & Deisenhofer, J. (1986) EMBO J. 3, 2445-2451], are delineated. The roles of several structural features in the preferential electron transfer along the A branch are discussed. Two bound detergent molecules of beta-octyl glucoside have been located near BA and BB. The environment of the carotenoid, C, that is present in RCs from Rb. sphaeroides 2.4.1 consists largely of aromatic residues of the M subunit. A role of BB in the triplet energy transfer from D to C and the reason for the preferential ease of removal of BB from the RC is proposed.
- University of California, Los Angeles 90024.
Organizational Affiliation: