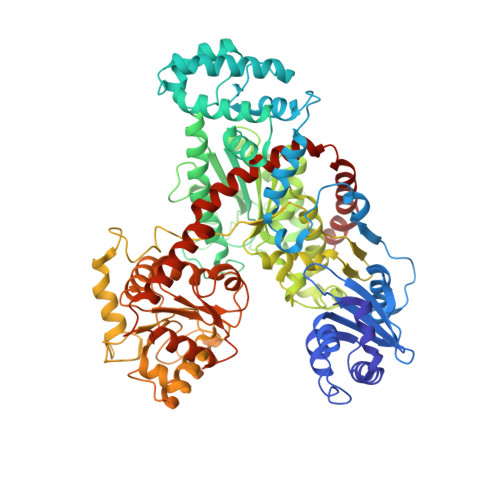
The Crystal Structure of Nitrosomonas europaea Sucrose Synthase Reveals Critical Conformational Changes and Insights into Sucrose Metabolism in Prokaryotes.
Wu, R., Asencion Diez, M.D., Figueroa, C.M., Machtey, M., Iglesias, A.A., Ballicora, M.A., Liu, D.(2015) J Bacteriol 197: 2734-2746
- PubMed: 26013491
- DOI: https://doi.org/10.1128/JB.00110-15
- Primary Citation of Related Structures:
4RBN - PubMed Abstract:
In this paper we report the first crystal structure of a prokaryotic sucrose synthase from the nonphotosynthetic bacterium Nitrosomonas europaea. The obtained structure was in an open form, whereas the only other available structure, from the plant Arabidopsis thaliana, was in a closed conformation. Comparative structural analysis revealed a "hinge-latch" combination, which is critical to transition between the open and closed forms of the enzyme. The N. europaea sucrose synthase shares the same fold as the GT-B family of the retaining glycosyltransferases. In addition, a triad of conserved homologous catalytic residues in the family was shown to be functionally critical in the N. europaea sucrose synthase (Arg567, Lys572, and Glu663). This implies that sucrose synthase shares not only a common origin with the GT-B family but also a similar catalytic mechanism. The enzyme preferred transferring glucose from ADP-glucose rather than UDP-glucose like the eukaryotic counterparts. This predicts that these prokaryotic organisms have a different sucrose metabolic scenario from plants. Nucleotide preference determines where the glucose moiety is targeted after sucrose is degraded. We obtained biochemical and structural evidence of sucrose metabolism in nonphotosynthetic bacteria. Until now, only sucrose synthases from photosynthetic organisms have been characterized. Here, we provide the crystal structure of the sucrose synthase from the chemolithoautotroph N. europaea. The structure supported that the enzyme functions with an open/close induced fit mechanism. The enzyme prefers as the substrate adenine-based nucleotides rather than uridine-based like the eukaryotic counterparts, implying a strong connection between sucrose and glycogen metabolism in these bacteria. Mutagenesis data showed that the catalytic mechanism must be conserved not only in sucrose synthases but also in all other retaining GT-B glycosyltransferases.
- Department of Chemistry and Biochemistry, Loyola University Chicago, Chicago, Illinois, USA.
Organizational Affiliation: